Saiba como você pode visualizar, modelar e entender seus projetos ambientais criando e definindo plumas de contaminação usando as ferramentas geoestatísticas robustas da Extensão de contaminação para Leapfrog Works a fim de obter estimativas transparentes e defensáveis da massa e localização de contaminantes em seus projetos.
Este webinar abordará:
- Uma introdução à Extensão de contaminação para Leapfrog Works.
- O fluxo de trabalho de extensão Contaminants — com uma breve visão geral de:
- Preparação e visualização de dados
- Análise exploratória de dados e criação de domínios
- Estimativa
- Validação do modelo
- Geração de relatórios
- Sessão de perguntas e respostas ao vivo.
Visão geral
Palestrantes
Aaron Hollingsworth
Especialista em soluções para clientes da Seequent
Steve Law
Líder técnico sênior – Sequente
Duração
55 minutos
Veja mais vídeos sob demanda
VídeosSaiba mais sobre a solução para engenharia civil da Seequent
Saiba maisTranscrição do vídeo
[00:00:00.320]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />So hello everyone<encoded_tag_open />/v<encoded_tag_closed /><!– wpml:html_fragment </p> –>
<p>[00:00:01.153]<br />
and welcome to today’s webinar</p>
<p>[00:00:02.400]<br />
on the Contaminants Extension for Leapfrog Works.</p>
<p>[00:00:05.990]<br />
Today we’ll be looking at the demonstration</p>
<p>[00:00:07.530]<br />
of the Contaminant Extension</p>
<p>[00:00:09.083]<br />
with time at the end of the webinar for your questions.</p>
<p>[00:00:12.460]<br />
My name is Aaron and I’m a Customer Solution Specialist</p>
<p>[00:00:15.540]<br />
here at Seequent, based in Brisbane office</p>
<p>[00:00:17.980]<br />
and I’m joined today by Steve Law</p>
<p>[00:00:19.710]<br />
our Senior Technical Lead for Geology.</p>
<p>[00:00:21.850]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />Hello, I’m Steve<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:00:22.850]<br />
and I’m a Senior Technical Lead</p>
<p>[00:00:24.520]<br />
and I’m based here in (mumbles).</p>
<p>[00:00:30.180]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />Today we’ll first go through<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:00:31.280]<br />
a bit of background on Seequent</p>
<p>[00:00:32.680]<br />
and the Contaminants Extension.</p>
<p>[00:00:34.900]<br />
I’ll then hand over to Steve who will take us through</p>
<p>[00:00:37.680]<br />
the Contaminant Extension in Leapfrog Works itself.</p>
<p>[00:00:41.610]<br />
After that we’ll go through any questions that anyone has.</p>
<p>[00:00:44.240]<br />
So please if you do have any at any time,</p>
<p>[00:00:46.470]<br />
put them in here at the question or the chat window</p>
<p>[00:00:48.890]<br />
and go to webinar and we’ll get to them.</p>
<p>[00:00:53.604]<br />
There’s some background on Seequent.</p>
<p>[00:00:56.070]<br />
We are a global leader</p>
<p>[00:00:57.230]<br />
in the development of visual data science software</p>
<p>[00:00:59.470]<br />
and collaborative technologies.</p>
<p>[00:01:01.560]<br />
With our software you can turn complex data</p>
<p>[00:01:03.610]<br />
into geological understanding, providing timely insights</p>
<p>[00:01:07.220]<br />
and give decision makers confidence.</p>
<p>[00:01:10.270]<br />
At Seequent, our mission is to enable our customers</p>
<p>[00:01:12.640]<br />
to make better decisions about their earth</p>
<p>[00:01:14.590]<br />
and environmental challenges.</p>
<p>[00:01:18.430]<br />
The origins of Seequent go back to the 1990s</p>
<p>[00:01:21.570]<br />
when some smart people</p>
<p>[00:01:22.680]<br />
at the University of Canterbury in New Zealand,</p>
<p>[00:01:24.720]<br />
created the fast radio basis functions</p>
<p>[00:01:27.170]<br />
for efficiently generating 3D surfaces from point data.</p>
<p>[00:01:31.460]<br />
One of the applications that was seen for this</p>
<p>[00:01:33.300]<br />
was a better way to build geological modeling.</p>
<p>[00:01:36.080]<br />
And so in 2004, the company ARANZ Geo was founded</p>
<p>[00:01:39.490]<br />
and the Leapfrog software was first released</p>
<p>[00:01:41.680]<br />
to the mining industry.</p>
<p>[00:01:44.250]<br />
Since then the company has grown and evolved</p>
<p>[00:01:46.580]<br />
entering different industries</p>
<p>[00:01:48.240]<br />
such as geothermal energy, environmental and civil.</p>
<p>[00:01:52.960]<br />
And in 2017, ARANZ Geo rebranded to Seequent.</p>
<p>[00:01:58.000]<br />
In 2018, Seequent acquired Geosoft</p>
<p>[00:02:00.670]<br />
the developers of Geophysical Software</p>
<p>[00:02:02.520]<br />
such as a Oasis montaj.</p>
<p>[00:02:04.520]<br />
And then in 2019, acquired GeoStudio,</p>
<p>[00:02:07.710]<br />
the developers of Geotechnical Analysis Software.</p>
<p>[00:02:11.930]<br />
During this time Seequent has continued developing</p>
<p>[00:02:15.630]<br />
our 3D geological modeling in Leapfrog</p>
<p>[00:02:18.620]<br />
and collaboration tools such as Central.</p>
<p>[00:02:22.480]<br />
Earlier this year,</p>
<p>[00:02:23.313]<br />
Seequent was acquired by Bentley Systems</p>
<p>[00:02:25.940]<br />
and has become a Bentley company</p>
<p>[00:02:27.433]<br />
which opens us up to even more possibilities</p>
<p>[00:02:30.430]<br />
for future development for geoscience solutions.</p>
<p>[00:02:36.310]<br />
Seequent have office globally</p>
<p>[00:02:37.990]<br />
with our head office in Christchurch, New Zealand.</p>
<p>[00:02:40.890]<br />
And more locally we have offices in Australia,</p>
<p>[00:02:43.550]<br />
in Perth and in Brisbane.</p>
<p>[00:02:48.240]<br />
Like I mentioned Seequent’s involved</p>
<p>[00:02:49.970]<br />
in number of different industries</p>
<p>[00:02:52.140]<br />
covering things from a contaminant modeling,</p>
<p>[00:02:55.550]<br />
road and toll construction,</p>
<p>[00:02:57.360]<br />
groundwater detection and management,</p>
<p>[00:02:59.460]<br />
geothermal exploration, resource evaluation</p>
<p>[00:03:02.560]<br />
and so much more.</p>
<p>[00:03:08.210]<br />
Today we’re looking at contaminant</p>
<p>[00:03:10.253]<br />
or contaminated and environmental projects.</p>
<p>[00:03:13.340]<br />
Seequent provides solutions across the life</p>
<p>[00:03:15.650]<br />
of contaminated site projects</p>
<p>[00:03:17.060]<br />
from the initial desk study phase</p>
<p>[00:03:19.140]<br />
to the site investigation</p>
<p>[00:03:20.800]<br />
through to the remediation, design and execution.</p>
<p>[00:03:25.070]<br />
These solutions include our geophysics software</p>
<p>[00:03:27.320]<br />
such as Oasis montaj and VOXI.</p>
<p>[00:03:29.930]<br />
Our 3D visualization and geological modeling</p>
<p>[00:03:32.490]<br />
and contaminant modeling in Leapfrog Works</p>
<p>[00:03:35.460]<br />
as well as flow and transport analysis in GeoStudio.</p>
<p>[00:03:39.710]<br />
Across all of this we have Seequent Central</p>
<p>[00:03:42.100]<br />
a cloud-based model management and collaboration tool</p>
<p>[00:03:45.320]<br />
for the life of the projects.</p>
<p>[00:03:48.090]<br />
Like I said, today we’re specifically looking</p>
<p>[00:03:50.220]<br />
at the Contaminants Extension in Leapfrog Works.</p>
<p>[00:03:56.040]<br />
So what is the Contaminants Extension?</p>
<p>[00:03:58.670]<br />
The Contaminants Extension</p>
<p>[00:03:59.790]<br />
is an optional module for Leapfrog works</p>
<p>[00:04:01.660]<br />
to enable spatial modeling of numeric data</p>
<p>[00:04:04.540]<br />
for defining the concentration, distribution</p>
<p>[00:04:07.330]<br />
and mass of the institute contamination at a site.</p>
<p>[00:04:11.910]<br />
The Seequent Contaminants Extension provides</p>
<p>[00:04:14.670]<br />
a transparent and auditable data-driven interpretation.</p>
<p>[00:04:19.900]<br />
It provides interactive visual tools</p>
<p>[00:04:22.430]<br />
with the extension that makes geostatistics</p>
<p>[00:04:24.760]<br />
accessible for all geoscientists</p>
<p>[00:04:27.610]<br />
ensuring that you and your teams</p>
<p>[00:04:29.120]<br />
can work the best with your data.</p>
<p>[00:04:32.600]<br />
It has built-in report tables</p>
<p>[00:04:34.730]<br />
and cross-section evaluations</p>
<p>[00:04:36.270]<br />
that are dynamically linked</p>
<p>[00:04:37.620]<br />
to your plume models and your data.</p>
<p>[00:04:41.350]<br />
So they will update automatically</p>
<p>[00:04:43.430]<br />
as the investigation and the project evolves in new day</p>
<p>[00:04:47.010]<br />
that comes into your Leapfrog project.</p>
<p>[00:04:50.320]<br />
It also means there’s an interrogatable estimation tool</p>
<p>[00:04:54.730]<br />
that provides easy access to the details</p>
<p>[00:04:58.260]<br />
for how you’ve created your informed decisions.</p>
<p>[00:05:02.595]<br />
In short, it allows you to take known data for your project</p>
<p>[00:05:07.070]<br />
and analyze it with spatial variability</p>
<p>[00:05:09.500]<br />
of the concentrations for your data</p>
<p>[00:05:12.390]<br />
and it helps you define your projects.</p>
<p>[00:05:20.860]<br />
So that’s what it is.</p>
<p>[00:05:22.410]<br />
Why did we develop it? And why is it useful?</p>
<p>[00:05:25.800]<br />
Well, we did it because we wanted to provide</p>
<p>[00:05:27.840]<br />
these accessible best practice solution tools</p>
<p>[00:05:30.690]<br />
for you to model your contamination plumes.</p>
<p>[00:05:34.290]<br />
We wanted to make characterizing contaminated land</p>
<p>[00:05:36.810]<br />
in groundwater, in a rigorous and auditable way</p>
<p>[00:05:39.730]<br />
that combines 3D dynamic geological Models</p>
<p>[00:05:42.700]<br />
with these best practice geostatistical methods.</p>
<p>[00:05:47.500]<br />
The ultimate goal of estimation</p>
<p>[00:05:48.970]<br />
is to combine the qualitative geological interpretation</p>
<p>[00:05:52.310]<br />
of your project.</p>
<p>[00:05:53.680]<br />
So your geological models</p>
<p>[00:05:55.270]<br />
with the sparsely sampled quantitative data</p>
<p>[00:05:58.450]<br />
to create spatial predictions</p>
<p>[00:06:00.130]<br />
for the distribution of that contaminant plan.</p>
<p>[00:06:09.140]<br />
The Contaminants Extension</p>
<p>[00:06:10.610]<br />
coupled with Seequent Central provides a wholistic way</p>
<p>[00:06:13.877]<br />
for the engineer and modeler for a project</p>
<p>[00:06:16.780]<br />
to effectively understand</p>
<p>[00:06:18.580]<br />
and importantly communicate the site conditions</p>
<p>[00:06:21.490]<br />
with both internal and external stakeholders</p>
<p>[00:06:24.310]<br />
in a collaborative environment.</p>
<p>[00:06:27.090]<br />
The graphic here shows an overview of the flow</p>
<p>[00:06:29.610]<br />
from Leapfrog to Central as part of the solution.</p>
<p>[00:06:34.350]<br />
Starting at the base here and moving up</p>
<p>[00:06:37.690]<br />
we can see that with your investigation data models</p>
<p>[00:06:40.982]<br />
and estimations can be created by the modeler</p>
<p>[00:06:44.260]<br />
and the environmental engineer for instance.</p>
<p>[00:06:48.310]<br />
And using Central,</p>
<p>[00:06:49.600]<br />
they can share this and collaborate on this</p>
<p>[00:06:53.000]<br />
with internal team members such as managers,</p>
<p>[00:06:55.830]<br />
reviewers or other modelers</p>
<p>[00:06:58.360]<br />
to build up the understanding of the site</p>
<p>[00:07:02.040]<br />
or as part of the project review process.</p>
<p>[00:07:06.420]<br />
At the very top of this infographic here</p>
<p>[00:07:08.260]<br />
we can then see how we can use Central</p>
<p>[00:07:10.330]<br />
to engage with external stakeholders</p>
<p>[00:07:13.130]<br />
such as the clients or other contractors</p>
<p>[00:07:16.650]<br />
and you can bring them into the Central project again</p>
<p>[00:07:18.740]<br />
to communicate in an interactive and visual way.</p>
<p>[00:07:24.820]<br />
The Seequent solution for the contaminant projects then</p>
<p>[00:07:27.490]<br />
is all about knowing the impact</p>
<p>[00:07:29.490]<br />
of environmental contamination for your projects.</p>
<p>[00:07:32.880]<br />
Through first seeing the problem</p>
<p>[00:07:34.360]<br />
in a clear, intuitive and visual way.</p>
<p>[00:07:38.030]<br />
The picture is worth 1,000 words</p>
<p>[00:07:39.440]<br />
a 3D visualization of the site has to be worth a million.</p>
<p>[00:07:44.260]<br />
With a Contaminants Extension,</p>
<p>[00:07:45.810]<br />
it’s an interactive dynamic site model</p>
<p>[00:07:47.990]<br />
you can create and understand</p>
<p>[00:07:50.130]<br />
and aid in your analysis and your assumptions</p>
<p>[00:07:53.070]<br />
and recommendations for your reports.</p>
<p>[00:08:01.340]<br />
I’ll now hand over to Steve</p>
<p>[00:08:03.500]<br />
who will take us through Leapfrog</p>
<p>[00:08:05.540]<br />
and the Contaminants Extension.</p>
<p>[00:08:27.267]<br />
So we’ll just switch over from the slides here. (mumbles).</p>
<p>[00:08:38.840]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />Thanks very much Aaron<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:08:40.756]<br />
for that great introduction.</p>
<p>[00:08:42.290]<br />
So I’m going to go through the</p>
<p>[00:08:44.470]<br />
Contaminants Extension Workflow.</p>
<p>[00:08:48.830]<br />
The end result is a resource model</p>
<p>[00:08:50.820]<br />
and through the whole process we have a look at our data.</p>
<p>[00:08:53.770]<br />
We go through the mining,</p>
<p>[00:08:55.340]<br />
run through estimations and validate and report at the end.</p>
<p>[00:08:59.650]<br />
Throughout the whole process it’s not a linear</p>
<p>[00:09:01.960]<br />
it can be a cyclical process</p>
<p>[00:09:03.840]<br />
so that whilst we’re at any stage</p>
<p>[00:09:06.310]<br />
we can always go back to our geology</p>
<p>[00:09:08.480]<br />
and look at how things are relating to each other.</p>
<p>[00:09:15.580]<br />
We’re going to go through a couple of key steps</p>
<p>[00:09:18.950]<br />
that we need to consider whilst building up a plume model.</p>
<p>[00:09:24.070]<br />
So the first part is being well aware of our data</p>
<p>[00:09:27.960]<br />
and how we can visualize it very easily</p>
<p>[00:09:30.240]<br />
within Leapfrog Works.</p>
<p>[00:09:31.980]<br />
So we want to consider things,</p>
<p>[00:09:33.659]<br />
where is our data and how is it located especially?</p>
<p>[00:09:37.500]<br />
We have tools in-built that we can check</p>
<p>[00:09:39.640]<br />
and make sure our data is validated</p>
<p>[00:09:42.490]<br />
and we have really good visualization tools.</p>
<p>[00:09:47.360]<br />
So within this project we have three models,</p>
<p>[00:09:51.440]<br />
the first one is a geology model</p>
<p>[00:09:53.840]<br />
and you can see here that it’s got outwash</p>
<p>[00:09:59.580]<br />
and ice deposits until above a bedrock sequence.</p>
<p>[00:10:03.960]<br />
And within that we have a range of vertical boreholes</p>
<p>[00:10:07.210]<br />
and these have been sampled generally in isolated locations</p>
<p>[00:10:13.840]<br />
for a chlorine contaminant</p>
<p>[00:10:16.030]<br />
and that’s what we’re going to be evaluating.</p>
<p>[00:10:19.240]<br />
As well as the geology model,</p>
<p>[00:10:21.440]<br />
we have a water table model.</p>
<p>[00:10:24.870]<br />
And this has been divided into a saturated</p>
<p>[00:10:28.040]<br />
and made (mumbles) again above bedrock.</p>
<p>[00:10:31.740]<br />
So one of the first things we want to consider is,</p>
<p>[00:10:34.910]<br />
is there any specific way that we should domain our data?</p>
<p>[00:10:38.040]<br />
To ask some of them, is chlorine associated with pathology?</p>
<p>[00:10:42.960]<br />
Is it associated with water table or a combination of both?</p>
<p>[00:10:47.199]<br />
And we have some tools within Leapfrog Works</p>
<p>[00:10:49.460]<br />
that can show this.</p>
<p>[00:10:52.180]<br />
So up in the boreholes’ database here,</p>
<p>[00:10:55.010]<br />
I have a table with the contaminants</p>
<p>[00:11:00.320]<br />
and then I’ve got a geology table.</p>
<p>[00:11:02.370]<br />
We have a merge table function</p>
<p>[00:11:04.090]<br />
so I’ve been able to merge these together.</p>
<p>[00:11:06.370]<br />
And this enables me then to evaluate them</p>
<p>[00:11:10.700]<br />
using some statistical tools</p>
<p>[00:11:13.120]<br />
in this case I’m using a box plot.</p>
<p>[00:11:17.640]<br />
And so this was here as a box plot of the geology</p>
<p>[00:11:20.360]<br />
against the contaminant.</p>
<p>[00:11:22.610]<br />
And we can see here that that is telling me</p>
<p>[00:11:24.910]<br />
that the outwash and local ice to contact deposits</p>
<p>[00:11:28.310]<br />
and the two, have the highest concentration of chlorine.</p>
<p>[00:11:32.310]<br />
And then there’s a lot less amount</p>
<p>[00:11:34.350]<br />
in the outwash deposits and bedrock.</p>
<p>[00:11:38.830]<br />
We can also have a look at this as a table of statistics.</p>
<p>[00:11:48.536]<br />
Excuse me.</p>
<p>[00:11:58.460]<br />
An idea of…</p>
<p>[00:11:59.810]<br />
We have a little bit total concentration.</p>
<p>[00:12:02.200]<br />
I can see the actual values within each one.</p>
<p>[00:12:06.220]<br />
One of the key things here is</p>
<p>[00:12:07.510]<br />
looking at the number of samples</p>
<p>[00:12:09.010]<br />
so we have only two samples within the bedrock.</p>
<p>[00:12:12.060]<br />
We’d identified that the outwash and ice deposits</p>
<p>[00:12:14.920]<br />
and the two had the greatest concentrations</p>
<p>[00:12:20.080]<br />
and we’ve got the most number of samples in those as well</p>
<p>[00:12:22.650]<br />
so we’ve got approximately 70 samples within the ice.</p>
<p>[00:12:27.310]<br />
And we’ve got the mains</p>
<p>[00:12:28.780]<br />
so we’ve got much higher means within ice</p>
<p>[00:12:32.660]<br />
and very low within the outwash deposits</p>
<p>[00:12:35.760]<br />
and again quite low</p>
<p>[00:12:36.970]<br />
but there’s only two samples within bedrock.</p>
<p>[00:12:41.280]<br />
So in this instance there’s no clear</p>
<p>[00:12:46.220]<br />
if we have a look at (mumbles)</p>
<p>[00:12:49.820]<br />
water table, we can see that both are saturated</p>
<p>[00:12:54.180]<br />
and the vital sign has significant concentrations</p>
<p>[00:12:59.030]<br />
and there’s not one particularly higher than the other.</p>
<p>[00:13:01.610]<br />
Saturated is a little bit more,</p>
<p>[00:13:03.600]<br />
but it’s not obviously completely different.</p>
<p>[00:13:08.900]<br />
So the way we’ll consider this then</p>
<p>[00:13:10.870]<br />
is I’m just going to separate a single domain</p>
<p>[00:13:13.940]<br />
that’s above bedrock and not taking into account</p>
<p>[00:13:16.790]<br />
whether it’s saturated or (mumbles).</p>
<p>[00:13:20.030]<br />
In this way, I have a geological model then</p>
<p>[00:13:23.370]<br />
that is just domained.</p>
<p>[00:13:37.140]<br />
So, once we’ve decided on</p>
<p>[00:13:38.630]<br />
how we’re going to domain our data,</p>
<p>[00:13:40.620]<br />
then we start using the Contaminants Extension information</p>
<p>[00:13:43.990]<br />
and we can link it to our geology model</p>
<p>[00:13:46.040]<br />
right through to the resource estimate.</p>
<p>[00:13:48.060]<br />
And we can do exploratory data analysis</p>
<p>[00:13:50.400]<br />
specifically on the domain itself.</p>
<p>[00:13:53.635]<br />
So we have a look at the domain model.</p>
<p>[00:13:57.190]<br />
And in this case</p>
<p>[00:13:58.023]<br />
this has now been all of the mythologies about bedrock</p>
<p>[00:14:02.320]<br />
have been combined into a single domain</p>
<p>[00:14:03.660]<br />
which is the yellow one on the scene.</p>
<p>[00:14:06.370]<br />
And we can see that the majority of the samples</p>
<p>[00:14:08.147]<br />
are within that and there’s just the one isolated to</p>
<p>[00:14:12.140]<br />
below in the bedrock which we won’t be considering</p>
<p>[00:14:14.960]<br />
in this particular estimate.</p>
<p>[00:14:24.890]<br />
When we build up estimation,</p>
<p>[00:14:29.130]<br />
we have what’s termed as contaminants model folder.</p>
<p>[00:14:32.230]<br />
So when we activate the Contaminants Extension License,</p>
<p>[00:14:37.600]<br />
it displays up as a folder in your project tree</p>
<p>[00:14:41.380]<br />
and there’s a estimation sub folder</p>
<p>[00:14:43.347]<br />
and a block model’s folder.</p>
<p>[00:14:45.490]<br />
The estimation folder can be considered to be</p>
<p>[00:14:47.950]<br />
where you set up all your parameters.</p>
<p>[00:14:49.930]<br />
And then the block model is where we display</p>
<p>[00:14:52.500]<br />
all of our results</p>
<p>[00:14:53.440]<br />
and we also do all our reporting and validation</p>
<p>[00:14:56.140]<br />
from that point as well.</p>
<p>[00:14:58.520]<br />
So I’ve decided to do a title constraint traction</p>
<p>[00:15:02.410]<br />
of this chloride within the unconsolidated domain</p>
<p>[00:15:05.870]<br />
which is this yellow block here.</p>
<p>[00:15:08.010]<br />
So when we create a domain estimator,</p>
<p>[00:15:12.920]<br />
we just select any closed solid within our project</p>
<p>[00:15:17.040]<br />
and you can see here that</p>
<p>[00:15:17.890]<br />
this is the unconsolidated sediments</p>
<p>[00:15:21.430]<br />
within our domain model.</p>
<p>[00:15:24.160]<br />
And then the numeric values are coming directly</p>
<p>[00:15:26.940]<br />
from our essay table, contaminant intervals,</p>
<p>[00:15:30.681]<br />
total concentration.</p>
<p>[00:15:32.760]<br />
So we can choose any numeric column</p>
<p>[00:15:34.860]<br />
within any part of our database.</p>
<p>[00:15:36.860]<br />
And also if we were storing point data</p>
<p>[00:15:39.520]<br />
within the points folder over here,</p>
<p>[00:15:41.630]<br />
we would be able to access that as well.</p>
<p>[00:15:43.980]<br />
So it doesn’t have to be within drew holes .</p>
<p>[00:15:48.720]<br />
I’m not applying any transform at this stage</p>
<p>[00:15:51.680]<br />
and we also have the capability of doing compositing.</p>
<p>[00:15:55.340]<br />
This data set doesn’t render itself to compositing</p>
<p>[00:15:58.790]<br />
because it’s got isolated values within each holes</p>
<p>[00:16:02.640]<br />
really only got one or two samples</p>
<p>[00:16:05.628]<br />
compositing small for if we’ve got a</p>
<p>[00:16:08.080]<br />
continuous range of samples down the borehole,</p>
<p>[00:16:12.020]<br />
and we’d want to composite that into large intervals</p>
<p>[00:16:17.510]<br />
to reduce their variance.</p>
<p>[00:16:20.170]<br />
So within each estimation for each domain,</p>
<p>[00:16:22.730]<br />
we have this sub folder.</p>
<p>[00:16:24.520]<br />
And from there we can see the domain itself.</p>
<p>[00:16:28.280]<br />
So we can double check that that is the volume of interest</p>
<p>[00:16:33.030]<br />
and we can display the values within that domain.</p>
<p>[00:16:37.700]<br />
So they have become point values at this stage</p>
<p>[00:16:42.800]<br />
and you can label those</p>
<p>[00:16:44.820]<br />
and you can also do statistics on these.</p>
<p>[00:16:47.610]<br />
So these now become domain statistics.</p>
<p>[00:16:50.500]<br />
So by right clicking on the values,</p>
<p>[00:16:54.280]<br />
I get a histogram of those values there.</p>
<p>[00:16:58.910]<br />
As with all Leapfrog Works graphs,</p>
<p>[00:17:04.010]<br />
they are interacting with the 3D scene.</p>
<p>[00:17:07.130]<br />
So let’s just move this over here.</p>
<p>[00:17:12.920]<br />
If I wanted to find out where these core values were,</p>
<p>[00:17:17.590]<br />
I can just highlight the one that on the graph</p>
<p>[00:17:20.610]<br />
and that will show up in the same format.</p>
<p>[00:17:23.450]<br />
Let’s do these ones about 100, there they are there.</p>
<p>[00:17:27.350]<br />
So gives us then idea of whether or not</p>
<p>[00:17:30.160]<br />
we need to create sub domains.</p>
<p>[00:17:32.400]<br />
If all the high grades were focused a little bit more</p>
<p>[00:17:35.520]<br />
we might be able to subdivide them out.</p>
<p>[00:17:37.210]<br />
But in this case,</p>
<p>[00:17:38.630]<br />
we’re just going to treat everything in the one domain.</p>
<p>[00:17:44.870]<br />
The special models folder is where we can do variography</p>
<p>[00:17:49.990]<br />
which is a spatial analysis of the data</p>
<p>[00:17:52.670]<br />
to see whether there are any specific directions</p>
<p>[00:17:55.320]<br />
where the concentration grades might be aligned.</p>
<p>[00:18:02.980]<br />
I’ll open this one up.</p>
<p>[00:18:12.510]<br />
So the way the variography works</p>
<p>[00:18:14.320]<br />
is that we select the direction</p>
<p>[00:18:16.700]<br />
and then we model it in three directions.</p>
<p>[00:18:21.370]<br />
One with the greatest degree of continuity et cetera.</p>
<p>[00:18:25.160]<br />
This radio plot is a plane within this.</p>
<p>[00:18:29.390]<br />
We’re looking in a fairly horizontal plane here</p>
<p>[00:18:32.667]<br />
and you can see that we have…</p>
<p>[00:18:35.600]<br />
we can always see the ellipsoid that we’re working with</p>
<p>[00:18:38.900]<br />
so to see whether it’s in the like</p>
<p>[00:18:41.160]<br />
we wanted to make that a little bit flatter.</p>
<p>[00:18:43.250]<br />
We didn’t leave it in a 3D scene</p>
<p>[00:18:45.660]<br />
and it will update on the form here</p>
<p>[00:18:48.210]<br />
and the graphs will update.</p>
<p>[00:18:51.260]<br />
We can use this radial plot.</p>
<p>[00:18:54.520]<br />
This may suggest that this orientation</p>
<p>[00:18:57.210]<br />
has a little bit more continuity and we’ll re-adjust.</p>
<p>[00:19:02.520]<br />
This particular data set doesn’t create</p>
<p>[00:19:05.680]<br />
a very good experimental variogram</p>
<p>[00:19:08.130]<br />
so we do have some other options</p>
<p>[00:19:09.710]<br />
where we could use a log transform prior to that</p>
<p>[00:19:14.293]<br />
so here I’ve done a log transform on the values</p>
<p>[00:19:19.450]<br />
and that was simply done by at the very beginning.</p>
<p>[00:19:22.550]<br />
When I selected my samples</p>
<p>[00:19:24.256]<br />
I could apply a log transform in through here.</p>
<p>[00:19:28.630]<br />
This gets rid of some of the variability</p>
<p>[00:19:30.650]<br />
especially the range and extreme differences</p>
<p>[00:19:34.350]<br />
between very high values and very low values</p>
<p>[00:19:37.260]<br />
and this can help develop a better semi-variably model</p>
<p>[00:19:42.300]<br />
if we have a little bit this one.</p>
<p>[00:19:50.624]<br />
You can see here that the semivariogram</p>
<p>[00:19:53.240]<br />
is a little bit better structured than it was before.</p>
<p>[00:19:56.700]<br />
Again this data is quite sparse.</p>
<p>[00:19:58.760]<br />
So this is about the best we could do</p>
<p>[00:20:01.070]<br />
with this particular data set.</p>
<p>[00:20:04.090]<br />
We have the ability to pick a couple of range of structures,</p>
<p>[00:20:08.970]<br />
we have a nugget plus a potentially two structures</p>
<p>[00:20:12.370]<br />
but in this case I’ll just pick a nugget,</p>
<p>[00:20:15.360]<br />
a fairly large nugget and a single structure</p>
<p>[00:20:18.030]<br />
which is sufficient to model this.</p>
<p>[00:20:21.190]<br />
We can get an idea of that model.</p>
<p>[00:20:23.350]<br />
So this is in the…</p>
<p>[00:20:25.090]<br />
The range is quite large, 1200 to 1400 meters.</p>
<p>[00:20:29.920]<br />
And that’s because the data itself is quite widely spaced.</p>
<p>[00:20:33.620]<br />
And we’ve only got a very short range</p>
<p>[00:20:35.390]<br />
in the Z direction because we’re looking at a</p>
<p>[00:20:37.294]<br />
fairly constrained tabular body of potential contamination.</p>
<p>[00:20:44.340]<br />
The advantage to building the variography</p>
<p>[00:20:46.390]<br />
within the contaminants extension is that</p>
<p>[00:20:49.070]<br />
everything is self-contained,</p>
<p>[00:20:50.410]<br />
and once we hit the side button,</p>
<p>[00:20:52.830]<br />
then that variogram is all ready to use</p>
<p>[00:20:56.510]<br />
within the estimations itself.</p>
<p>[00:21:00.480]<br />
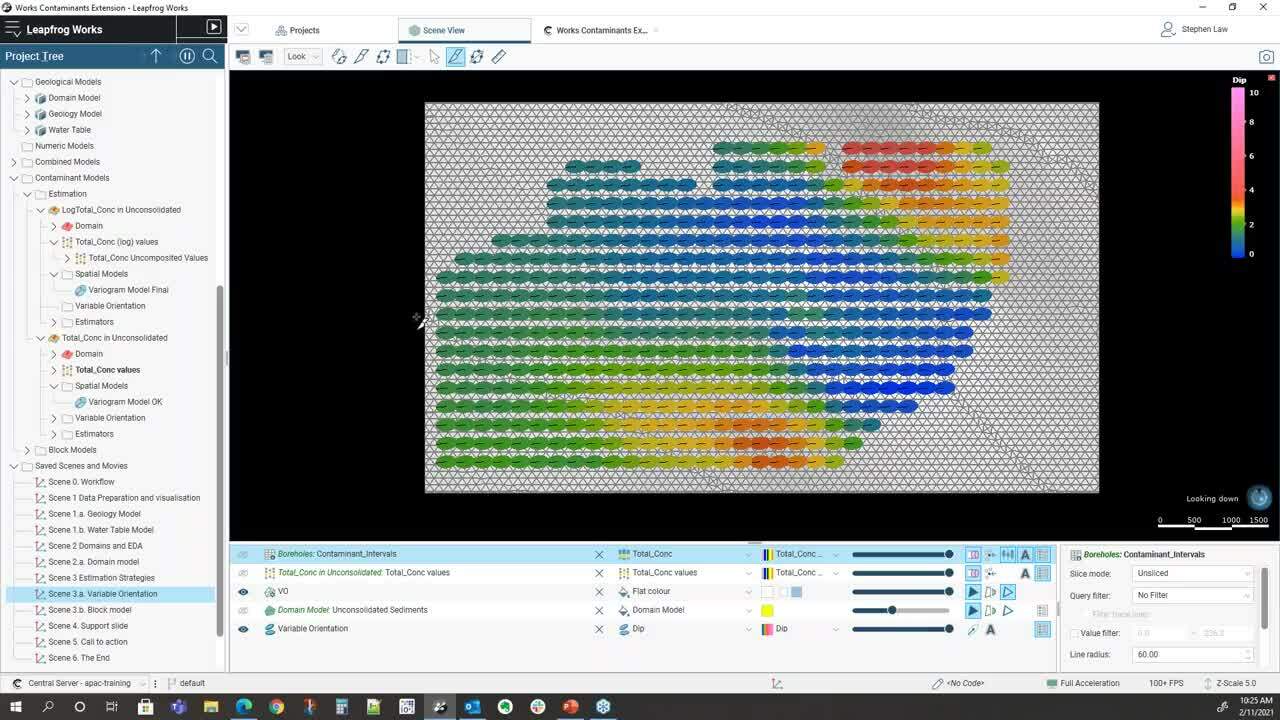
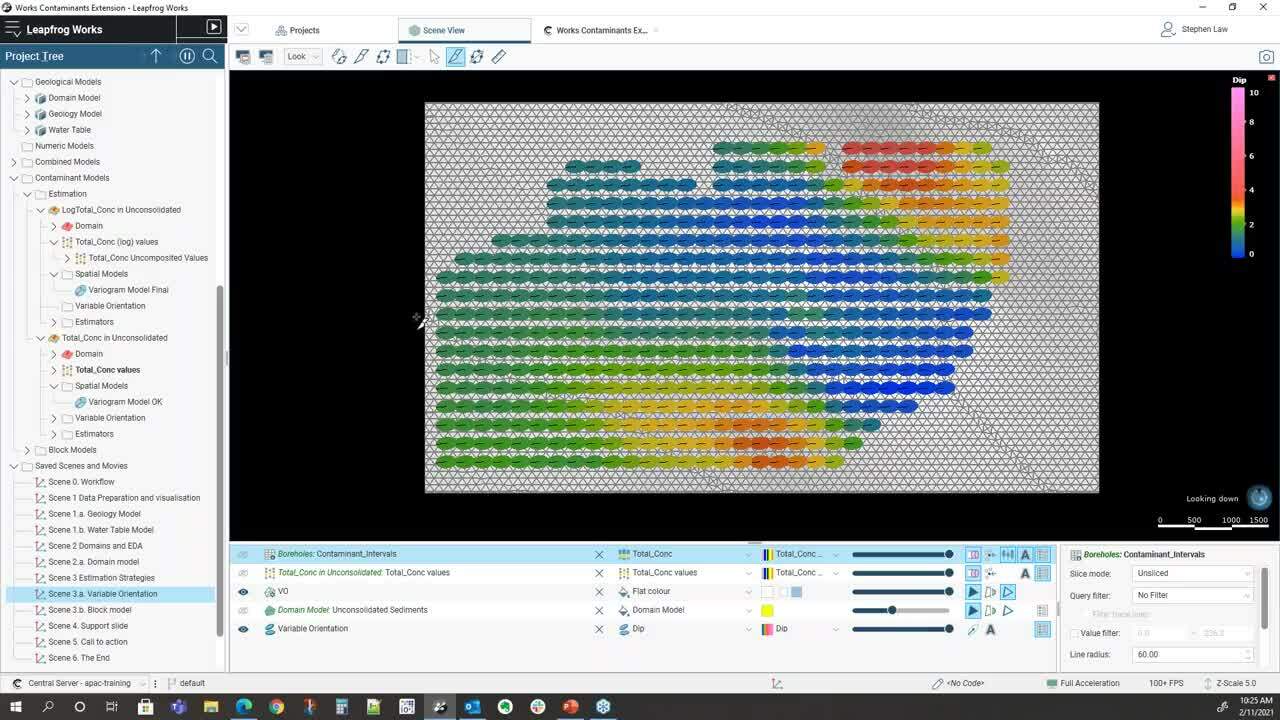
We also have a concept of variable orientation.</p>
<p>[00:21:04.750]<br />
Sometimes the design of the Contaminant</p>
<p>[00:21:10.520]<br />
isn’t purely tabular</p>
<p>[00:21:12.480]<br />
and there may be local irregularities</p>
<p>[00:21:14.810]<br />
within the surfaces.</p>
<p>[00:21:16.680]<br />
So we can use those surfaces</p>
<p>[00:21:18.450]<br />
to help guide the local orientation of the variogram</p>
<p>[00:21:22.750]<br />
when we’re doing sample selection.</p>
<p>[00:21:25.470]<br />
So just don’t want it here already.</p>
<p>[00:21:31.890]<br />
So the way we do that</p>
<p>[00:21:32.810]<br />
is that we set up a reference surface</p>
<p>[00:21:35.440]<br />
and in this case it’s the actual base</p>
<p>[00:21:37.570]<br />
over the top of the bedrock</p>
<p>[00:21:39.530]<br />
and we can see here that we’ve colored this by a dip.</p>
<p>[00:21:44.480]<br />
So we can see that we’ve got some varying dips</p>
<p>[00:21:47.670]<br />
that flat up in the middle</p>
<p>[00:21:49.070]<br />
and slightly more dips around the edges.</p>
<p>[00:21:51.830]<br />
If I can hop and look cross sections through there,</p>
<p>[00:22:03.450]<br />
we can see that what will happen</p>
<p>[00:22:06.610]<br />
is when we pick out samples,</p>
<p>[00:22:09.060]<br />
the ellipsoid will be orientated as local</p>
<p>[00:22:12.780]<br />
and so overall ellipsoid is in this general direction,</p>
<p>[00:22:16.010]<br />
but where it gets flat out,</p>
<p>[00:22:17.400]<br />
it will rotate in a flat orientation.</p>
<p>[00:22:21.570]<br />
The actual ratios and the variogram parameters themselves</p>
<p>[00:22:24.960]<br />
are still maintained.</p>
<p>[00:22:26.267]<br />
It’s just the orientation</p>
<p>[00:22:28.260]<br />
for when the samples are being selected.</p>
<p>[00:22:35.270]<br />
When we’re doing an estimation,</p>
<p>[00:22:36.730]<br />
we have to consider a few different things.</p>
<p>[00:22:39.330]<br />
So we need to understand what the stage our project is</p>
<p>[00:22:42.650]<br />
and this might help us work out</p>
<p>[00:22:44.520]<br />
which estimation method to use.</p>
<p>[00:22:46.550]<br />
If we’ve got early stage,</p>
<p>[00:22:48.190]<br />
we might not have very much data.</p>
<p>[00:22:49.910]<br />
We might not have sufficient data to get a variogram.</p>
<p>[00:22:52.740]<br />
So we will need to use either nearest neighbor</p>
<p>[00:22:55.510]<br />
or inverse distance type methods.</p>
<p>[00:22:59.476]<br />
There can be a lot of work to go into the search strategy.</p>
<p>[00:23:02.280]<br />
And that is how many samples are we going to use</p>
<p>[00:23:05.061]<br />
to define a local estimate.</p>
<p>[00:23:08.850]<br />
And then once we run our estimate,</p>
<p>[00:23:10.900]<br />
we need to validate it against the original data</p>
<p>[00:23:14.250]<br />
to make sure that we’re maintaining the same kind of means</p>
<p>[00:23:17.950]<br />
and then we want to report our results at the end.</p>
<p>[00:23:23.150]<br />
To set up the estimators,</p>
<p>[00:23:25.970]<br />
we work through this estimators sub folder here</p>
<p>[00:23:29.760]<br />
and if you right click</p>
<p>[00:23:30.920]<br />
you can go have inverse distance nearest neighbor kriging</p>
<p>[00:23:34.857]<br />
and we can do ordinary kriging, simple kriging</p>
<p>[00:23:38.010]<br />
or we can use the actual RBF estimator as well.</p>
<p>[00:23:41.520]<br />
Which can be quite good</p>
<p>[00:23:42.870]<br />
for when we haven’t got sufficient data to a variogram.</p>
<p>[00:23:47.110]<br />
It’s a global estimator.</p>
<p>[00:23:49.540]<br />
Using the same underlying algorithm</p>
<p>[00:23:51.470]<br />
that is used for when we krig the surfaces</p>
<p>[00:23:54.170]<br />
or the numeric models that can be run</p>
<p>[00:23:56.600]<br />
within Leapfrog Works itself.</p>
<p>[00:24:00.710]<br />
To set one up</p>
<p>[00:24:01.990]<br />
once I open we can edit that,</p>
<p>[00:24:04.650]<br />
we can apply clipping which is</p>
<p>[00:24:08.390]<br />
if we want to constrain the very high values</p>
<p>[00:24:11.010]<br />
and give them less influence,</p>
<p>[00:24:12.550]<br />
we can apply that on floor.</p>
<p>[00:24:15.090]<br />
In this case because I’ve actually looking at contaminants</p>
<p>[00:24:17.380]<br />
I don’t really want to cut them down initially</p>
<p>[00:24:19.580]<br />
so I will leave them unclicked.</p>
<p>[00:24:22.540]<br />
I’m just doing point kriging in this case</p>
<p>[00:24:24.710]<br />
so just I’m not using discretization at all.</p>
<p>[00:24:28.590]<br />
In some cases if I want to block kriging,</p>
<p>[00:24:30.460]<br />
then I would increase these discretization amounts.</p>
<p>[00:24:35.170]<br />
I’m using ordinary kriging.</p>
<p>[00:24:36.693]<br />
This is where we determine ordinary or simple.</p>
<p>[00:24:41.120]<br />
And this is the variogram.</p>
<p>[00:24:42.750]<br />
So you can pick.</p>
<p>[00:24:44.180]<br />
It will see the variogram within the folder.</p>
<p>[00:24:50.460]<br />
You set to a particular variogram</p>
<p>[00:24:53.090]<br />
and that’s more for making sure</p>
<p>[00:24:55.730]<br />
that it’s set to the correct orientation.</p>
<p>[00:24:58.240]<br />
And then it’ll automatically go to the maximum range</p>
<p>[00:25:01.100]<br />
of that variogram but you can modify these</p>
<p>[00:25:03.680]<br />
to be less than that or greater.</p>
<p>[00:25:05.790]<br />
Depends on the circumstances</p>
<p>[00:25:08.750]<br />
and often you’ll do a lot of testing</p>
<p>[00:25:11.230]<br />
and sensitivity analysis to determine</p>
<p>[00:25:13.760]<br />
the best scope for this one.</p>
<p>[00:25:18.400]<br />
And then we can choose</p>
<p>[00:25:19.890]<br />
minimum and maximum number of samples.</p>
<p>[00:25:22.610]<br />
And this effectively becomes</p>
<p>[00:25:24.280]<br />
our grade variable within the block model</p>
<p>[00:25:27.710]<br />
and we can store other variables as well.</p>
<p>[00:25:30.510]<br />
So if we’re using kriging,</p>
<p>[00:25:32.030]<br />
we can store things such as kriging variance,</p>
<p>[00:25:34.240]<br />
slope and regression</p>
<p>[00:25:35.820]<br />
which would go as to how good the estimate might be.</p>
<p>[00:25:39.430]<br />
We can store things such as average distance to samples</p>
<p>[00:25:42.290]<br />
and these may help us with classification</p>
<p>[00:25:44.810]<br />
criteria down the track.</p>
<p>[00:25:47.150]<br />
So I’ve created us an ordinary kriged estimator here</p>
<p>[00:25:52.640]<br />
and then I’ve also got one.</p>
<p>[00:25:54.270]<br />
This one is exactly the same,</p>
<p>[00:25:57.340]<br />
but in this case I’ve actually applied</p>
<p>[00:25:59.610]<br />
the variable orientation.</p>
<p>[00:26:01.230]<br />
So we’ve set up a variable orientation within the folder.</p>
<p>[00:26:04.190]<br />
It will be accessible in our parameter set up through here</p>
<p>[00:26:09.050]<br />
and I’ll give it a slightly different name.</p>
<p>[00:26:11.550]<br />
This one I will need to compare the kriging results</p>
<p>[00:26:17.310]<br />
with and without variable orientation apply.</p>
<p>[00:26:21.570]<br />
So we set up all of our different parameters</p>
<p>[00:26:24.250]<br />
in this estimation folder.</p>
<p>[00:26:27.360]<br />
And we are then able to create a block model</p>
<p>[00:26:32.690]<br />
to look at the results.</p>
<p>[00:26:40.300]<br />
Setting up a block model is quite simple</p>
<p>[00:26:43.440]<br />
so we can create a new block model</p>
<p>[00:26:45.900]<br />
and we can also import block models from other software.</p>
<p>[00:26:49.790]<br />
And when we’ve created one,</p>
<p>[00:26:51.990]<br />
we can edit it by opening it up.</p>
<p>[00:26:55.500]<br />
And in this case,</p>
<p>[00:26:56.333]<br />
we’ve got a block size of 80 by 80 meters</p>
<p>[00:26:58.720]<br />
in the X, Y direction and five in the Z</p>
<p>[00:27:02.260]<br />
so we’re looking at that fairly thin.</p>
<p>[00:27:04.270]<br />
And if we’re looking at it again very visual</p>
<p>[00:27:07.167]<br />
and we can see our block size.</p>
<p>[00:27:13.230]<br />
So this is 80 by 80 in the X Y direction here.</p>
<p>[00:27:18.400]<br />
And we can change these boundaries at any time.</p>
<p>[00:27:20.810]<br />
So once it’s been made,</p>
<p>[00:27:21.900]<br />
if we wanted to adjust it,</p>
<p>[00:27:23.533]<br />
I could just move the boundary up</p>
<p>[00:27:25.720]<br />
and it will automatically update the model.</p>
<p>[00:27:28.930]<br />
I want to do that now so it doesn’t rerun.</p>
<p>[00:27:31.820]<br />
A key concept when working with block models</p>
<p>[00:27:34.030]<br />
within the Contaminants Extension is this evaluations tab.</p>
<p>[00:27:38.260]<br />
So basically whatever we want to flag into the block model</p>
<p>[00:27:41.794]<br />
needs to come across from the left-hand side</p>
<p>[00:27:44.590]<br />
across to the right.</p>
<p>[00:27:46.040]<br />
So what I’m doing here</p>
<p>[00:27:47.090]<br />
is I’m flagging each block centroid with the domain model</p>
<p>[00:27:50.980]<br />
so that will flag it with whether it’s within that domain</p>
<p>[00:27:56.590]<br />
but we can also flag at geology.</p>
<p>[00:27:58.410]<br />
So this will put into whether it’s two</p>
<p>[00:28:00.770]<br />
or whether it’s bedrock</p>
<p>[00:28:02.610]<br />
and also avoid us saturated sides</p>
<p>[00:28:05.110]<br />
could be flagged as well.</p>
<p>[00:28:07.500]<br />
And then this is Y grade or contaminant variables here.</p>
<p>[00:28:12.870]<br />
So I’m just putting in all four that are generated</p>
<p>[00:28:17.200]<br />
so I’ve got an inverse distance,</p>
<p>[00:28:19.360]<br />
I’ve got two different krigings.</p>
<p>[00:28:21.734]<br />
And also the nearest neighbor.</p>
<p>[00:28:23.460]<br />
Nearest neighbor can be useful</p>
<p>[00:28:25.140]<br />
for helping with validation of the model.</p>
<p>[00:28:29.480]<br />
Again, any of these can be removed or added at one time.</p>
<p>[00:28:32.680]<br />
So if I did want to look at the RBF,</p>
<p>[00:28:35.070]<br />
I would just bring it across</p>
<p>[00:28:37.170]<br />
and then the model would rerun.</p>
<p>[00:28:41.200]<br />
So we can see here, the model has been generated.</p>
<p>[00:28:45.710]<br />
And one of the key things for validation of first</p>
<p>[00:28:49.120]<br />
is to actually look at the samples against the results.</p>
<p>[00:28:54.550]<br />
So if I just go into a cross section</p>
<p>[00:29:06.010]<br />
making sure that our color schemes</p>
<p>[00:29:08.760]<br />
forbides the data and the bot model are the same</p>
<p>[00:29:11.940]<br />
and we can import these colors</p>
<p>[00:29:13.580]<br />
so if we’ve got colors defined for our essays up here,</p>
<p>[00:29:23.952]<br />
we can export those colors</p>
<p>[00:29:25.640]<br />
like create a little file,</p>
<p>[00:29:27.360]<br />
and then we can go down to our block model</p>
<p>[00:29:29.660]<br />
and we can import those colors down here.</p>
<p>[00:29:34.420]<br />
So I did on this one, look at that colors import</p>
<p>[00:29:38.370]<br />
and then pick up that file.</p>
<p>[00:29:39.670]<br />
And that way we get exactly same color scheme.</p>
<p>[00:29:42.640]<br />
And what we’re hoping to see</p>
<p>[00:29:43.910]<br />
is a little we’ve got yellows</p>
<p>[00:29:46.610]<br />
we should hopefully have orange or yellows around,</p>
<p>[00:29:49.710]<br />
higher grades will be in the reds and purples</p>
<p>[00:29:53.610]<br />
and lower grades should show up.</p>
<p>[00:30:02.308]<br />
Of course we have a lot of data in here.</p>
<p>[00:30:06.074]<br />
There we go.</p>
<p>[00:30:06.907]<br />
So we can see some low grade points over here</p>
<p>[00:30:09.280]<br />
and that’s equating.</p>
<p>[00:30:11.170]<br />
We’ve got really good tools for looking at the details</p>
<p>[00:30:14.870]<br />
of the results on a block-by-block basis.</p>
<p>[00:30:17.720]<br />
If I highlighted this particular block here,</p>
<p>[00:30:21.250]<br />
we have this interrogate estimator function.</p>
<p>[00:30:26.130]<br />
And this is the estimator that it’s looking at.</p>
<p>[00:30:29.080]<br />
So like ordinary krig without variable orientation</p>
<p>[00:30:32.560]<br />
it’s found there’s many samples</p>
<p>[00:30:35.390]<br />
and depending on how we’ve set up our search,</p>
<p>[00:30:38.120]<br />
in this case it’s included all those samples.</p>
<p>[00:30:41.050]<br />
It’s showing us the kriging lights</p>
<p>[00:30:42.360]<br />
that have been applied and we can sort these</p>
<p>[00:30:45.177]<br />
and distances from the sample.</p>
<p>[00:30:48.660]<br />
So we can see the closest sample used was 175 meters away</p>
<p>[00:30:53.080]<br />
and the greatest one was over 1,000 meters away.</p>
<p>[00:30:56.200]<br />
But it’s got lights through there.</p>
<p>[00:31:00.470]<br />
Part of the kriging process</p>
<p>[00:31:01.730]<br />
can give negative kriging weights</p>
<p>[00:31:03.670]<br />
and that is not a problem</p>
<p>[00:31:04.850]<br />
unless we start to see negative grade values.</p>
<p>[00:31:07.770]<br />
In that case, you might need to change</p>
<p>[00:31:09.520]<br />
our sample selection a little bit</p>
<p>[00:31:12.660]<br />
to eradicate that influence.</p>
<p>[00:31:16.420]<br />
That’s just part of the kriging process.</p>
<p>[00:31:21.130]<br />
You can see also that when we do that we can…</p>
<p>[00:31:23.740]<br />
it shows the ellipsoid for that area.</p>
<p>[00:31:26.720]<br />
And if we filter the block model</p>
<p>[00:31:30.818]<br />
and I’ll just take away the ellipsoid for a second,</p>
<p>[00:31:36.180]<br />
it shows you the block and it shows you the samples</p>
<p>[00:31:39.113]<br />
that are being used.</p>
<p>[00:31:40.540]<br />
So you’ve got a nice spatial visualization</p>
<p>[00:31:44.000]<br />
of what samples are actually being used</p>
<p>[00:31:46.080]<br />
to estimate the grade within that box through there.</p>
<p>[00:31:48.947]<br />
And if you want to try something else,</p>
<p>[00:31:50.760]<br />
you can go directly to the estimator and modify.</p>
<p>[00:31:55.360]<br />
If I want to change this to three samples,</p>
<p>[00:31:58.070]<br />
reduce these chains to search</p>
<p>[00:32:00.600]<br />
and you can get see what the impacts could be. Okay.</p>
<p>[00:32:06.790]<br />
So apart from visualization</p>
<p>[00:32:08.910]<br />
and looking at on a block-by-block basis,</p>
<p>[00:32:12.620]<br />
we can also run swath plots.</p>
<p>[00:32:18.780]<br />
So within the block model itself,</p>
<p>[00:32:20.870]<br />
we can create a new swath plot.</p>
<p>[00:32:23.210]<br />
And the beauty of this is that</p>
<p>[00:32:25.200]<br />
they are embedded within the block model</p>
<p>[00:32:27.090]<br />
so that when we update that data later on,</p>
<p>[00:32:29.750]<br />
the swath plots will update as well</p>
<p>[00:32:32.270]<br />
and the same goes for reports.</p>
<p>[00:32:34.060]<br />
If we build a report on the model</p>
<p>[00:32:36.170]<br />
then if we change the data,</p>
<p>[00:32:37.650]<br />
then the reports will have tables.</p>
<p>[00:32:40.409]<br />
So we’ll have a look at a swath</p>
<p>[00:32:42.610]<br />
that’s already been prepared</p>
<p>[00:32:44.030]<br />
and you can store any number of swaths within the model.</p>
<p>[00:32:49.330]<br />
So what this one is doing</p>
<p>[00:32:50.530]<br />
is it’s looking at the inverse distance.</p>
<p>[00:32:53.470]<br />
It’s got the nearest neighbor which is this gray</p>
<p>[00:32:56.170]<br />
and then we’ve got two different types of kriging.</p>
<p>[00:32:59.950]<br />
So we can see straight away.</p>
<p>[00:33:01.320]<br />
And it’s showing us in the X, Y and Z directions</p>
<p>[00:33:05.810]<br />
relative to the axis of the block model.</p>
<p>[00:33:08.830]<br />
So if we look in the X direction here,</p>
<p>[00:33:10.640]<br />
we can see that all three estimators</p>
<p>[00:33:13.647]<br />
are fairly close to each other</p>
<p>[00:33:15.780]<br />
and what the red line is is the actual original sample data.</p>
<p>[00:33:20.450]<br />
So these are the sample values</p>
<p>[00:33:22.250]<br />
and what we generally do want to see</p>
<p>[00:33:24.920]<br />
is that the estimators tend to bisect</p>
<p>[00:33:27.110]<br />
whether we have these highs and lows,</p>
<p>[00:33:29.280]<br />
the estimated values we should run through the middle.</p>
<p>[00:33:32.600]<br />
Also the estimator should follow the trend</p>
<p>[00:33:35.570]<br />
across the data.</p>
<p>[00:33:37.600]<br />
The nearest neighbor can be used as a validation tool.</p>
<p>[00:33:42.090]<br />
So if one of these estimators</p>
<p>[00:33:43.880]<br />
for instance was sitting quite high</p>
<p>[00:33:46.330]<br />
up here above their sniper line</p>
<p>[00:33:48.810]<br />
they would suggest that potentially where I over estimated</p>
<p>[00:33:50.850]<br />
and there might be some problems with our parameters</p>
<p>[00:33:54.040]<br />
so we would want to change that.</p>
<p>[00:33:56.810]<br />
Again we can export, we can copy this data</p>
<p>[00:34:00.537]<br />
and put it into a spreadsheet.</p>
<p>[00:34:02.160]<br />
Sometimes people have very specific formats</p>
<p>[00:34:06.650]<br />
they like to do their swath plots in.</p>
<p>[00:34:08.200]<br />
So that will bring all the data out into Excel.</p>
<p>[00:34:10.690]<br />
So you can do that yourself</p>
<p>[00:34:12.640]<br />
or we can just take the image</p>
<p>[00:34:14.830]<br />
and paste it directly into our report.</p>
<p>[00:34:21.780]<br />
Okay. If we want to report on our data,</p>
<p>[00:34:24.950]<br />
we create the report in the block model.</p>
<p>[00:34:28.400]<br />
And in this case, I’ve got just the basic report.</p>
<p>[00:34:33.710]<br />
That’s looking at the geology and the water table</p>
<p>[00:34:38.020]<br />
and it’s giving us the totals and the concentration.</p>
<p>[00:34:41.560]<br />
And this is by having to select these categorical columns</p>
<p>[00:34:45.400]<br />
and then we have our grade or value columns.</p>
<p>[00:34:48.030]<br />
So we need at least one categorical column</p>
<p>[00:34:50.820]<br />
and one to recreate a report</p>
<p>[00:34:53.310]<br />
but you can have as many different categories</p>
<p>[00:34:56.150]<br />
as you like within there.</p>
<p>[00:34:58.160]<br />
And these are also act like a pivot table almost</p>
<p>[00:35:01.030]<br />
so if I want it to show geology first</p>
<p>[00:35:04.370]<br />
and then want a table,</p>
<p>[00:35:05.760]<br />
I can split it up through there like that.</p>
<p>[00:35:08.220]<br />
We can apply a cutoff or we can have none as well.</p>
<p>[00:35:12.740]<br />
Another advantage within the block models (mumbles)</p>
<p>[00:35:20.090]<br />
is that we can create calculations and filters</p>
<p>[00:35:26.150]<br />
and we can get quite complex with these.</p>
<p>[00:35:30.040]<br />
We have a whole series of syntax helpers</p>
<p>[00:35:34.060]<br />
which are over here on the right.</p>
<p>[00:35:36.590]<br />
And then in middle here,</p>
<p>[00:35:37.750]<br />
it shows us all of our parameters</p>
<p>[00:35:39.620]<br />
that are available within the block model</p>
<p>[00:35:41.720]<br />
to use within a calculation.</p>
<p>[00:35:45.630]<br />
It’s also a nice little summary of the values</p>
<p>[00:35:48.800]<br />
within each estimator.</p>
<p>[00:35:52.030]<br />
Now you saw here for instance,</p>
<p>[00:35:53.900]<br />
I can see that I’ve got a couple of negatives</p>
<p>[00:35:56.797]<br />
in this ordinary krig without the variable orientation.</p>
<p>[00:36:00.270]<br />
So I’d want to have a look at those</p>
<p>[00:36:01.760]<br />
to see where they are and whether I need to remove them</p>
<p>[00:36:04.920]<br />
or do something about them.</p>
<p>[00:36:07.660]<br />
If I just unpin that for a second</p>
<p>[00:36:10.600]<br />
we can see, we can get quite complex</p>
<p>[00:36:13.020]<br />
we’ve got lots of if statements we can embed them.</p>
<p>[00:36:16.500]<br />
So what’s happening with this one</p>
<p>[00:36:18.060]<br />
is I’m trying to define porosity</p>
<p>[00:36:20.380]<br />
based on the rock type within the model.</p>
<p>[00:36:24.240]<br />
So I’ve defined, if we go back to here,</p>
<p>[00:36:28.740]<br />
I can look at the geology.</p>
<p>[00:36:30.820]<br />
So the geology is being coded within to the model.</p>
<p>[00:36:34.300]<br />
And if I stick on that block you can see here</p>
<p>[00:36:36.470]<br />
the geology model is till and the water table is saturated.</p>
<p>[00:36:41.060]<br />
So I can use any of those components (mumbles)</p>
<p>[00:36:48.723]<br />
so stop saying, “If the domain model is still,</p>
<p>[00:36:51.740]<br />
I’m going to assign a plus to the 8.25.”</p>
<p>[00:36:54.320]<br />
And that’s a fairly simple calculation</p>
<p>[00:36:56.300]<br />
but you can put in as many complex calculations as you wish.</p>
<p>[00:37:00.300]<br />
This one title concentration is saying that</p>
<p>[00:37:02.720]<br />
it must be within unconsolidated sediments.</p>
<p>[00:37:06.420]<br />
And what this one is doing is saying,</p>
<p>[00:37:07.960]<br />
what happens if I’ve got some blocks</p>
<p>[00:37:09.580]<br />
that haven’t been given a value.</p>
<p>[00:37:12.140]<br />
So I can say that if there is no value,</p>
<p>[00:37:15.059]<br />
I’m going to assign a very low one</p>
<p>[00:37:17.570]<br />
but otherwise use the value if it’s greater than zero,</p>
<p>[00:37:21.270]<br />
use the value that’s there.</p>
<p>[00:37:22.510]<br />
So that’s one way of getting rid of negatives</p>
<p>[00:37:24.440]<br />
if you know that there’s only a</p>
<p>[00:37:26.330]<br />
couple of isolated ones there.</p>
<p>[00:37:30.680]<br />
And then this one is an average distance.</p>
<p>[00:37:32.530]<br />
So I store the average distance within the model.</p>
<p>[00:37:35.980]<br />
So I’m using this as a guide for classification.</p>
<p>[00:37:39.260]<br />
So stop saying if the average distance</p>
<p>[00:37:40.940]<br />
to the samples is less than 500 meters,</p>
<p>[00:37:43.500]<br />
I’ll make it measured less than 1,000 indicated</p>
<p>[00:37:46.621]<br />
less than 1500.</p>
<p>[00:37:48.970]<br />
The beauty of any calculation, numeric or categorical</p>
<p>[00:37:52.630]<br />
is that it’s available instantly to visualize.</p>
<p>[00:37:55.810]<br />
So we can visualize this confidence category.</p>
<p>[00:37:59.410]<br />
So you can see that there,</p>
<p>[00:38:00.610]<br />
I’ve just got indicated in third</p>
<p>[00:38:03.380]<br />
and a little bit of measured there as well.</p>
<p>[00:38:06.800]<br />
Let’s split that out. (mumbles)</p>
<p>[00:38:10.980]<br />
So we can see here,</p>
<p>[00:38:12.360]<br />
that’s based on the data and the data spacing.</p>
<p>[00:38:17.050]<br />
We might not use those values directly to classify</p>
<p>[00:38:19.860]<br />
but we can use them as a visual guide</p>
<p>[00:38:22.240]<br />
if we wanted to draw shapes to classify certain areas.</p>
<p>[00:38:28.580]<br />
So once we’ve done that kind of thing</p>
<p>[00:38:31.100]<br />
we can build another report based of that calculation.</p>
<p>[00:38:36.460]<br />
So in this case we’re using that calculated field</p>
<p>[00:38:40.060]<br />
as part of that categorical information.</p>
<p>[00:38:43.920]<br />
And so now we’ve got classification,</p>
<p>[00:38:46.180]<br />
the geology and the water table in the report.</p>
<p>[00:38:49.630]<br />
And again that’s saved automatically and it can be exported.</p>
<p>[00:38:53.890]<br />
We can copy it into an Excel Spreadsheet</p>
<p>[00:38:57.790]<br />
or you can just export it as an Excel Spreadsheet directly.</p>
<p>[00:39:05.930]<br />
Okay. So that’s the main usage</p>
<p>[00:39:10.240]<br />
of the Contaminants Extension.</p>
<p>[00:39:12.160]<br />
And I’ll just pass right back again to Aaron to finish it.</p>
<p>[00:39:16.290]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />Fantastic. Thanks Steve.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:39:20.160]<br />
We have time now for questions as well.</p>
<p>[00:39:23.670]<br />
So if you have them already,</p>
<p>[00:39:25.360]<br />
please feel free to put a question in the question box</p>
<p>[00:39:28.330]<br />
or you can raise your hand as well</p>
<p>[00:39:30.320]<br />
and we can unmute you</p>
<p>[00:39:31.670]<br />
and you can verbally ask the question as well.</p>
<p>[00:39:35.650]<br />
While we give people a chance to do that,</p>
<p>[00:39:39.289]<br />
beyond today’s meeting there’s always support for Leapfrog</p>
<p>[00:39:43.360]<br />
and the Contamination Extension</p>
<p>[00:39:44.880]<br />
by contacting us at [email protected]</p>
<p>[00:39:48.910]<br />
or by giving our support line a phone call.</p>
<p>[00:39:52.480]<br />
After today’s session as well,</p>
<p>[00:39:54.852]<br />
we’d love to give the extension of Leapfrog a go.</p>
<p>[00:39:59.620]<br />
We have a trial available</p>
<p>[00:40:00.790]<br />
that you can sign up for on our website</p>
<p>[00:40:02.480]<br />
on the Leapfrog Works products page</p>
<p>[00:40:04.450]<br />
which you can see the URL therefore.</p>
<p>[00:40:06.690]<br />
We’ll also be sending an email out after this webinar</p>
<p>[00:40:10.710]<br />
with the recording for today</p>
<p>[00:40:12.410]<br />
and also a link for a four-part technical series</p>
<p>[00:40:16.180]<br />
on the Contamination Extension</p>
<p>[00:40:17.840]<br />
which will take you through step-by-step</p>
<p>[00:40:20.330]<br />
how to build the model that Steve has shown you</p>
<p>[00:40:23.190]<br />
from scratch and the process behind that.</p>
<p>[00:40:27.440]<br />
And includes the data there as well.</p>
<p>[00:40:28.830]<br />
So that’s a fantastic resource we’ll be sending to you.</p>
<p>[00:40:31.860]<br />
We also have online learning content</p>
<p>[00:40:34.490]<br />
at my.seequent.com/learning.</p>
<p>[00:40:37.500]<br />
And we also have the</p>
<p>[00:40:38.830]<br />
Leapfrog Works and Contaminants Extension help pages</p>
<p>[00:40:41.910]<br />
will look out at them later.</p>
<p>[00:40:43.400]<br />
And that’s a really useful resource</p>
<p>[00:40:44.850]<br />
if you have any questions.</p>
<p>[00:40:51.770]<br />
So yeah like I said, if you have questions,</p>
<p>[00:40:54.510]<br />
please put them up or raise your hand.</p>
<p>[00:40:57.170]<br />
One of the questions that we have so far is</p>
<p>[00:41:01.392]<br />
so often contaminants sources</p>
<p>[00:41:03.840]<br />
are at the surface propagating down</p>
<p>[00:41:05.910]<br />
into the soil or groundwater</p>
<p>[00:41:09.872]<br />
and how would you go about modeling a change</p>
<p>[00:41:12.800]<br />
in the directional trend</p>
<p>[00:41:14.240]<br />
I guess of the plume going from vertical</p>
<p>[00:41:16.620]<br />
as it propagates down to spreading out horizontally.</p>
<p>[00:41:20.075]<br />
I guess once it’s an aquifer or an aquatar.</p>
<p>[00:41:24.710]<br />
So can you answer that for us please Steve?</p>
<p>[00:41:27.230]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />Yep so everything about the Contaminants Extension<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:41:30.750]<br />
is all about how we set up our domains</p>
<p>[00:41:33.360]<br />
at the very beginning.</p>
<p>[00:41:34.770]<br />
So it does need a physical three-dimensional representation</p>
<p>[00:41:39.056]<br />
of each domain.</p>
<p>[00:41:40.690]<br />
So the way we would do that would</p>
<p>[00:41:42.310]<br />
we would need acquifer surface and the soil.</p>
<p>[00:41:46.414]<br />
We would have one domain and say that</p>
<p>[00:41:49.390]<br />
then you’d model your aquifer as a horizontal layer.</p>
<p>[00:41:54.410]<br />
So you would set up two estimation folders,</p>
<p>[00:41:57.120]<br />
one for the soil, one for the aquifer.</p>
<p>[00:41:59.410]<br />
And then within the soil one,</p>
<p>[00:42:00.630]<br />
you could make your orientation vertical</p>
<p>[00:42:03.420]<br />
and within the aquifer you can change it to horizontal.</p>
<p>[00:42:06.850]<br />
And then you get…</p>
<p>[00:42:07.770]<br />
You’re able to join these things together</p>
<p>[00:42:10.120]<br />
using the calculation so that you can visualize</p>
<p>[00:42:13.240]<br />
both domains together in the block model.</p>
<p>[00:42:16.200]<br />
So you only need one block model</p>
<p>[00:42:17.920]<br />
but you can have multiple domains</p>
<p>[00:42:19.890]<br />
which will get twined together.</p>
<p>[00:42:21.950]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />Fantastic. Thank you Steve.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:42:24.770]<br />
Another question here is,</p>
<p>[00:42:26.600]<br />
were any of those calculations</p>
<p>[00:42:28.180]<br />
automatically populated by Leapfrog</p>
<p>[00:42:30.270]<br />
or did you make each of them?</p>
<p>[00:42:32.040]<br />
If you made them, just to be clear,</p>
<p>[00:42:33.530]<br />
did you make them to remove the negative values</p>
<p>[00:42:35.960]<br />
generated by the kriging?</p>
<p>[00:42:38.045]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />No, they’re not generated automatically.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:42:40.530]<br />
So I created all of those calculations</p>
<p>[00:42:43.450]<br />
and you don’t have to force removal of the negatives.</p>
<p>[00:42:48.510]<br />
It really comes down to first</p>
<p>[00:42:50.860]<br />
realizing that negative values exist.</p>
<p>[00:42:53.420]<br />
And then we would look at them in the 3D scene</p>
<p>[00:42:55.450]<br />
to see where they are.</p>
<p>[00:42:56.820]<br />
Now, if they’re only the isolated block around the edges</p>
<p>[00:43:00.260]<br />
quite a bit away from your data,</p>
<p>[00:43:03.010]<br />
then they might not be significant.</p>
<p>[00:43:05.060]<br />
So then you could use the calculation to zero them out.</p>
<p>[00:43:09.690]<br />
Sometimes you get a situation</p>
<p>[00:43:11.260]<br />
where you can get negative grades</p>
<p>[00:43:13.410]<br />
right within the main part of your data.</p>
<p>[00:43:16.387]<br />
And this is the thing called a kriging et cetera.</p>
<p>[00:43:21.763]<br />
and the kriging process can cause this.</p>
<p>[00:43:24.550]<br />
So in that case you would have to</p>
<p>[00:43:26.100]<br />
change your search selection.</p>
<p>[00:43:28.510]<br />
So you might have to decrease the maximum number of samples.</p>
<p>[00:43:31.750]<br />
You might need to tweak the variogram a little bit</p>
<p>[00:43:34.340]<br />
and then you should be able to get</p>
<p>[00:43:35.830]<br />
reduce the impact of negative weights</p>
<p>[00:43:38.219]<br />
So that you won’t get negative grades.</p>
<p>[00:43:41.450]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />To add to that first part of the question as well.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:43:43.942]<br />
Once you have created your calculations,</p>
<p>[00:43:47.230]<br />
you can actually go in and export them out</p>
<p>[00:43:51.760]<br />
to use in other projects as well.</p>
<p>[00:43:53.410]<br />
So kind of… Where are we?</p>
<p>[00:43:58.490]<br />
Calculations are cool.</p>
<p>[00:43:59.860]<br />
So calculations and filters.</p>
<p>[00:44:01.490]<br />
So in this calculations tab here,</p>
<p>[00:44:03.780]<br />
you can see we have this option here</p>
<p>[00:44:05.490]<br />
for importing or indeed for exporting here as well.</p>
<p>[00:44:08.840]<br />
So once you set it at once,</p>
<p>[00:44:10.440]<br />
you can reuse that in other projects.</p>
<p>[00:44:12.620]<br />
So it’s very easy to transfer these across projects</p>
<p>[00:44:16.260]<br />
maybe change some of the variable names.</p>
<p>[00:44:18.260]<br />
If you need to, it’s different under different projects.</p>
<p>[00:44:20.967]<br />
And it’s once you’ve set at once</p>
<p>[00:44:22.410]<br />
it’s very easy to use those again</p>
<p>[00:44:24.167]<br />
and that’s a files, it’s a calculation file</p>
<p>[00:44:26.380]<br />
that you can send to other people in your company as well.</p>
<p>[00:44:28.390]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />So if you don’t have the same variable names,<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:44:31.830]<br />
what it does,</p>
<p>[00:44:32.663]<br />
it just highlights them with a little red,</p>
<p>[00:44:34.430]<br />
underline and then you just replace it</p>
<p>[00:44:36.880]<br />
with the correct variable name.</p>
<p>[00:44:38.340]<br />
So yeah easy to set things up</p>
<p>[00:44:41.890]<br />
especially if you’re using the same format</p>
<p>[00:44:43.545]<br />
of block model over a whole project area</p>
<p>[00:44:46.400]<br />
for different block models.</p>
<p>[00:44:49.020]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />Fantastic. And yeah as a reminder,<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:44:51.230]<br />
if you do have questions,</p>
<p>[00:44:52.150]<br />
please put them in the chat or raise your hand.</p>
<p>[00:44:55.140]<br />
We have another question here.</p>
<p>[00:44:56.220]<br />
Can you run the steps to include the 95 percentile?</p>
<p>[00:45:01.562]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />Not directly,<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:45:04.331]<br />
actually we can get…</p>
<p>[00:45:07.090]<br />
We do do log probability plots.</p>
<p>[00:45:09.600]<br />
So (mumbles).</p>
<p>[00:45:13.350]<br />
So if I go back up to this one here yet</p>
<p>[00:45:24.680]<br />
so my daily statistics of strike first up</p>
<p>[00:45:27.940]<br />
get a histogram,</p>
<p>[00:45:30.169]<br />
but I can change this to a low probability plot.</p>
<p>[00:45:32.710]<br />
So then I’ll be able to see where the 95th percentile is.</p>
<p>[00:45:36.890]<br />
Is down there.</p>
<p>[00:45:38.570]<br />
So the 95th percentile is up through there like that.</p>
<p>[00:45:42.622]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />Awesome. Thanks Steve.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:45:45.850]<br />
So can you please show</p>
<p>[00:45:48.310]<br />
the contamination plume advance over time?</p>
<p>[00:45:50.790]<br />
And can you show the database with the time element column?</p>
<p>[00:45:54.990]<br />
So we don’t have any time or…</p>
<p>[00:45:58.534]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />Sorry.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:46:00.184]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />monitoring data set up for this project<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:46:03.321]<br />
but you could certainly do that.</p>
<p>[00:46:05.140]<br />
So you could just set up your first one,</p>
<p>[00:46:07.555]<br />
make a copy of it and then change the data to the new,</p>
<p>[00:46:10.730]<br />
but the next monitoring event.</p>
<p>[00:46:11.920]<br />
So if each monitoring event,</p>
<p>[00:46:13.654]<br />
you’re just using that separate data</p>
<p>[00:46:16.400]<br />
in the same estimations, in the same block models</p>
<p>[00:46:20.900]<br />
and also in with the contaminants kit.</p>
<p>[00:46:23.250]<br />
If you go into the points folder,</p>
<p>[00:46:25.335]<br />
you do have this option for</p>
<p>[00:46:26.900]<br />
import time dependent points as well.</p>
<p>[00:46:28.830]<br />
So you can import a point cloud</p>
<p>[00:46:30.070]<br />
that has the time data or attribute to it.</p>
<p>[00:46:33.370]<br />
You can actually filter that.</p>
<p>[00:46:34.390]<br />
So visually you can use a filter</p>
<p>[00:46:36.040]<br />
to see the data at a certain point in time</p>
<p>[00:46:38.340]<br />
but you could then actually</p>
<p>[00:46:40.050]<br />
if you wanted to, you could set up a workflow</p>
<p>[00:46:41.960]<br />
where you set filters up</p>
<p>[00:46:43.550]<br />
to use that point data in your block model</p>
<p>[00:46:46.630]<br />
and just filter it based on time as well.</p>
<p>[00:46:49.650]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />So when you create a estimator,<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:46:53.550]<br />
so this one here,</p>
<p>[00:46:59.390]<br />
you can see here you can apply a filter to your data.</p>
<p>[00:47:02.420]<br />
So yes, if you divided up</p>
<p>[00:47:05.030]<br />
created pre filters based on the time,</p>
<p>[00:47:07.660]<br />
you can run a series of block models.</p>
<p>[00:47:13.090]<br />
Well actually you could probably do it</p>
<p>[00:47:14.700]<br />
within the single block model.</p>
<p>[00:47:16.090]<br />
So you would just set up</p>
<p>[00:47:17.440]<br />
a different estimator for each time period,</p>
<p>[00:47:20.020]<br />
estimate them all and put them all,</p>
<p>[00:47:21.810]<br />
evaluate them all into the same block model</p>
<p>[00:47:24.260]<br />
and then you could kind of look at them and how it changes.</p>
<p>[00:47:28.070]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />Yeah. Awesome.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:47:29.640]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />That’d be a few different ways<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:47:30.790]<br />
of trying to do that.</p>
<p>[00:47:35.420]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />We have a question here<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:47:36.770]<br />
which is, when should you use the different</p>
<p>[00:47:41.750]<br />
or when should you use the different estimation methods</p>
<p>[00:47:45.190]<br />
or I guess?</p>
<p>[00:47:46.478]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />Guess so. It really comes down<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:47:48.860]<br />
to personal choice a little bit</p>
<p>[00:47:51.810]<br />
but generally, we do need a certain degree</p>
<p>[00:47:56.270]<br />
of the amount of data to do kriging</p>
<p>[00:47:58.590]<br />
so that we can get a variable variogram</p>
<p>[00:48:01.420]<br />
So early stages,</p>
<p>[00:48:02.670]<br />
we might only be able to do inverse distance</p>
<p>[00:48:06.510]<br />
whether it’s inverse distance squared or cubed et cetera.</p>
<p>[00:48:10.300]<br />
It’s again personal preference.</p>
<p>[00:48:13.517]<br />
Generally though, if we wanted to get</p>
<p>[00:48:15.280]<br />
quite a localized estimate,</p>
<p>[00:48:17.780]<br />
we may use inverse distance cubed,</p>
<p>[00:48:19.860]<br />
a more general one the inverse distance squared.</p>
<p>[00:48:23.580]<br />
And now if we can start to get a variogram out</p>
<p>[00:48:25.960]<br />
it’s always best to use kriging if possible</p>
<p>[00:48:28.777]<br />
it does tend.</p>
<p>[00:48:29.970]<br />
You find that, especially with sparse data,</p>
<p>[00:48:32.760]<br />
you won’t get too much,</p>
<p>[00:48:34.270]<br />
the results shouldn’t be too different from each other</p>
<p>[00:48:36.930]<br />
but kriging does tend to work with it better.</p>
<p>[00:48:41.150]<br />
The RBF, I don’t use it much myself</p>
<p>[00:48:44.173]<br />
but it can be quite useful to get early stage global idea</p>
<p>[00:48:49.590]<br />
of how things are going.</p>
<p>[00:48:50.790]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />Yeah. So when you have sparse<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:48:51.847]<br />
so not much data can be useful in those initial stages</p>
<p>[00:48:55.120]<br />
maybe as pilot or desk study phase or something.</p>
<p>[00:48:57.930]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />Yeah. Yeah.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:48:58.871]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />For some case (mumbles).<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:49:02.110]<br />
Another question here,</p>
<p>[00:49:03.020]<br />
can you please explain the hard and soft boundaries</p>
<p>[00:49:05.450]<br />
that appeared in the box just before</p>
<p>[00:49:08.280]<br />
so in that initial estimation and this one here.</p>
<p>[00:49:13.550]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />So what this is,<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:49:15.190]<br />
is that in this case it’s not</p>
<p>[00:49:17.408]<br />
or that sort of relevant.</p>
<p>[00:49:18.990]<br />
There are only two samples outside our domain.</p>
<p>[00:49:21.930]<br />
So if I bring back up my domain boundary,</p>
<p>[00:49:25.900]<br />
let’s do that, okay.</p>
<p>[00:49:31.090]<br />
So hard and soft basically</p>
<p>[00:49:32.982]<br />
is what happens around the boundary of the domain itself.</p>
<p>[00:49:37.950]<br />
How do I treat the samples?</p>
<p>[00:49:40.110]<br />
So a hard boundary in 99% of the time,</p>
<p>[00:49:43.720]<br />
you will use a hard boundary.</p>
<p>[00:49:45.430]<br />
Is that you saying,</p>
<p>[00:49:47.180]<br />
basically I’m only looking at samples</p>
<p>[00:49:48.930]<br />
within the domain itself,</p>
<p>[00:49:50.300]<br />
and I don’t care about anything outside it.</p>
<p>[00:49:53.630]<br />
The instance where you may use a soft boundary</p>
<p>[00:49:56.030]<br />
is if there’s a good directional change,</p>
<p>[00:49:59.660]<br />
say the concentration can’t be limited directly</p>
<p>[00:50:02.630]<br />
to one of the main or another.</p>
<p>[00:50:03.463]<br />
And there’s a good directional change across that boundary.</p>
<p>[00:50:07.200]<br />
By using a soft boundary, we can define,</p>
<p>[00:50:11.222]<br />
we could say 20 meters.</p>
<p>[00:50:13.600]<br />
And what that would do is we’ll look at for samples,</p>
<p>[00:50:16.660]<br />
20 meters beyond the boundary in all directions.</p>
<p>[00:50:19.650]<br />
And we’ll use those as part of the sample selection.</p>
<p>[00:50:23.350]<br />
You still only estimate within the domain,</p>
<p>[00:50:27.420]<br />
but around the edges you use samples</p>
<p>[00:50:29.860]<br />
that are just slightly outside.</p>
<p>[00:50:31.730]<br />
So that’s the main difference between hard and soft.</p>
<p>[00:50:35.090]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />And I guess going back<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:50:36.180]<br />
to a question and an answer</p>
<p>[00:50:37.330]<br />
that was before around using multiple domains</p>
<p>[00:50:40.260]<br />
to kind of handle the vertical and then horizontal</p>
<p>[00:50:42.900]<br />
would it be fair to say</p>
<p>[00:50:43.733]<br />
that maybe you’d use the soft boundary</p>
<p>[00:50:45.700]<br />
in that first one to accommodate the values near</p>
<p>[00:50:50.130]<br />
really that domain boundary into the horizontal?</p>
<p>[00:50:52.290]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />That’s right.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:50:53.250]<br />
And I mean, one of the beauties of a Leapfrog Works,</p>
<p>[00:50:57.960]<br />
the Contaminants et cetera,</p>
<p>[00:50:59.210]<br />
is that it’s so easy to test things.</p>
<p>[00:51:02.020]<br />
So once you’ve got one of these folders set up</p>
<p>[00:51:06.060]<br />
if you want to try something a little bit different,</p>
<p>[00:51:08.420]<br />
you just copy the entire folder</p>
<p>[00:51:11.470]<br />
and you might pick a slightly different domain</p>
<p>[00:51:14.630]<br />
or if you’re going to do it on a different element,</p>
<p>[00:51:16.910]<br />
choose it there.</p>
<p>[00:51:18.310]<br />
And let’s just copy. That’s fine.</p>
<p>[00:51:20.360]<br />
And it pre copies basically any very variables</p>
<p>[00:51:22.923]<br />
that you might have done or you estimated is a set up.</p>
<p>[00:51:25.720]<br />
And then you can just go back into that one and say,</p>
<p>[00:51:28.940]<br />
I want to try something that’s little bit different</p>
<p>[00:51:31.237]<br />
with this kriging set up.</p>
<p>[00:51:33.150]<br />
So you might try a different distance,</p>
<p>[00:51:35.710]<br />
some different samples et cetera.</p>
<p>[00:51:38.050]<br />
So then you would just run them,</p>
<p>[00:51:39.645]<br />
evaluate that into your bottle</p>
<p>[00:51:41.740]<br />
and look at it on a swath plot</p>
<p>[00:51:43.760]<br />
to see what the difference was.</p>
<p>[00:51:45.820]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />Awesome. Thanks Steve.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:51:48.370]<br />
I think you touched on maybe this question a little bit,</p>
<p>[00:51:51.040]<br />
but is there an output or evaluation or really…</p>
<p>[00:51:56.150]<br />
How can you show uncertainty of the block model?</p>
<p>[00:51:58.760]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />Okay. So there are lots of different ways<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:52:01.650]<br />
of trying to look at uncertainty.</p>
<p>[00:52:03.743]<br />
The simplest way is just looking at data spacing.</p>
<p>[00:52:07.469]<br />
And that’s a little bit what we’ve tried to do</p>
<p>[00:52:10.240]<br />
with the calculation field before.</p>
<p>[00:52:13.860]<br />
So I open up my calculations again.</p>
<p>[00:52:16.550]<br />
So we stored the average distance to the samples</p>
<p>[00:52:20.430]<br />
and that was done.</p>
<p>[00:52:23.690]<br />
I’m here in the outputs</p>
<p>[00:52:25.460]<br />
so I stored the average distance sample.</p>
<p>[00:52:27.530]<br />
You can also store the minimum distance</p>
<p>[00:52:29.720]<br />
to the closest sample.</p>
<p>[00:52:31.600]<br />
And then again by making up</p>
<p>[00:52:33.960]<br />
some kind of a calculation like this,</p>
<p>[00:52:38.130]<br />
if that’s not working, then I can change.</p>
<p>[00:52:40.370]<br />
And I could change that to 800, 1200 et cetera</p>
<p>[00:52:44.240]<br />
until it sort of makes sense.</p>
<p>[00:52:46.210]<br />
So we can use (mumbles).</p>
<p>[00:52:50.040]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />Yeah.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:52:50.873]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />We can use distance buffers,<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:52:52.060]<br />
like let’s add on</p>
<p>[00:52:53.900]<br />
Contaminants Extension functionality within works</p>
<p>[00:52:56.530]<br />
and that might help you as well</p>
<p>[00:52:58.440]<br />
decide on what kind of values to use</p>
<p>[00:53:00.630]<br />
within these calculations.</p>
<p>[00:53:02.300]<br />
Classification, if you’re using kriging,</p>
<p>[00:53:06.046]<br />
if you have used directly kriging variance</p>
<p>[00:53:09.270]<br />
as it often quite nicely to use spacing as well.</p>
<p>[00:53:12.980]<br />
So there’s lots of different ways,</p>
<p>[00:53:15.220]<br />
but in more of the LT user.</p>
<p>[00:53:17.520]<br />
<encoded_tag_open />v Aaron<encoded_tag_closed />Fantastic. Well thank you so much Steve.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:53:20.317]<br />
We’re coming to the end of…</p>
<p>[00:53:22.107]<br />
We had a lot of time now.</p>
<p>[00:53:23.830]<br />
Thank you, everyone who joined</p>
<p>[00:53:25.470]<br />
and particularly if you asked a question.</p>
<p>[00:53:27.902]<br />
If we didn’t get to your question</p>
<p>[00:53:29.800]<br />
or if you think of one after today’s session,</p>
<p>[00:53:32.360]<br />
then please feel free to reach out</p>
<p>[00:53:35.990]<br />
at the [email protected].</p>
<p>[00:53:38.100]<br />
And if there are anything we couldn’t get though today,</p>
<p>[00:53:40.480]<br />
we’ll also reach out directly to people.</p>
<p>[00:53:44.720]<br />
As I said, you will be receiving a link after today</p>
<p>[00:53:49.450]<br />
with the recording as well as</p>
<p>[00:53:51.183]<br />
the link for that technical series,</p>
<p>[00:53:53.700]<br />
which is a really good resource to go through yourself</p>
<p>[00:53:56.960]<br />
using the Contaminant Extension.</p>
<p>[00:53:59.160]<br />
And if you don’t already have access to it,</p>
<p>[00:54:00.790]<br />
we can give you a trial of it as well.</p>
<p>[00:54:03.410]<br />
So you can give it a go.</p>
<p>[00:54:07.840]<br />
Thank you so much for joining today’s webinar.</p>
<p>[00:54:09.960]<br />
We really hope you enjoyed it</p>
<p>[00:54:11.450]<br />
and have a fantastic rest of your day.</p>
<wpml_invalid_tag original=”PHA+” />[00:54:13.590]<br />
<encoded_tag_open />v Steve<encoded_tag_closed />Thank you very much.<encoded_tag_open />/v<encoded_tag_closed />