Permítanos mostrarle cómo Leapfrog Edge puede simplificar sus estimaciones de recursos con enlaces dinámicos y visualización en 3D, al poner la geología en el centro de nuestra estimación. También presentaremos un flujo de trabajo dinámico sobre el grado de grosor.
Generalidades
Oradores
Peter Oshust
Geólogo sénior de Desarrollo de Negocio, Seequent
Duración
39 min

Ver más videos bajo demanda.
VideosObtenga más información acerca de la solución para la minería de Seequent.
Más informaciónTranscripción del video
[00:00:00.899]<br />
(gentle music)<!– wpml:html_fragment </p> –>
<p>[00:00:10.870]<br />
<encoded_tag_open />v Peter<encoded_tag_closed />Good morning or good afternoon everyone<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:00:14.078]<br />
from wherever you’re attending.</p>
<p>[00:00:14.911]<br />
I’m a professional geologist</p>
<p>[00:00:17.462]<br />
with a few decades of experience</p>
<p>[00:00:18.470]<br />
in mineral exploration and mining.</p>
<p>[00:00:20.160]<br />
And I’ve focused mainly on long-term</p>
<p>[00:00:23.760]<br />
mineral resource estimates in a variety of commodities,</p>
<p>[00:00:27.810]<br />
diamonds, base metals, copper, nickel, precious metals</p>
<p>[00:00:32.540]<br />
and PGEs and through a variety of deposit styles</p>
<p>[00:00:36.950]<br />
in North and South America and Asia.</p>
<p>[00:00:40.260]<br />
The bulk of my experience</p>
<p>[00:00:41.920]<br />
was spent at the Ekati Diamond Mine,</p>
<p>[00:00:44.490]<br />
where I contributed to a team that did the resource updates</p>
<p>[00:00:48.476]<br />
on 10-kimberlite pipes up there at the time.</p>
<p>[00:00:52.610]<br />
And I was with Wood, formerly Amec Foster Wheeler</p>
<p>[00:00:56.610]<br />
for nine years in the Mining $ Metals Consulting group.</p>
<p>[00:01:00.900]<br />
I’ve been at Seequent now since October, 2018,</p>
<p>[00:01:03.360]<br />
so just over two years</p>
<p>[00:01:06.512]<br />
and I am on the technical team</p>
<p>[00:01:10.112]<br />
supporting business development training</p>
<p>[00:01:12.699]<br />
and providing technical support.</p>
<p>[00:01:13.790]<br />
And I think a few of you</p>
<p>[00:01:15.886]<br />
will probably have communicated with me in that respect.</p>
<p>[00:01:19.870]<br />
And I do focus on the Leapfrog Edge</p>
<p>[00:01:24.150]<br />
resource estimation tool in Geo.</p>
<p>[00:01:28.780]<br />
So we’re going to cover</p>
<p>[00:01:31.870]<br />
basically the estimation workflow in Edge.</p>
<p>[00:01:35.170]<br />
So we’ll start with exploratory data analysis</p>
<p>[00:01:37.650]<br />
and compositing, we’ll define a few estimators.</p>
<p>[00:01:41.330]<br />
We’ll create a rotated sub-blocked block model.</p>
<p>[00:01:43.947]<br />
And this is kind of the trick for doing</p>
<p>[00:01:45.940]<br />
the grade-thickness calculation in Edge.</p>
<p>[00:01:48.960]<br />
We’ll evaluate the estimators, validate the results.</p>
<p>[00:01:52.260]<br />
Not thoroughly, but we will do a couple of checks</p>
<p>[00:01:55.449]<br />
and then we’ll compose and evaluate</p>
<p>[00:01:56.810]<br />
our grade-thickness calculations</p>
<p>[00:01:59.020]<br />
and review the results.</p>
<p>[00:02:00.670]<br />
And then of course,</p>
<p>[00:02:02.240]<br />
our work is intended for another audience</p>
<p>[00:02:05.350]<br />
and I’m picturing that the rotated</p>
<p>[00:02:08.523]<br />
grade-thickness model will go to</p>
<p>[00:02:12.060]<br />
engineers who will re-block it</p>
<p>[00:02:14.160]<br />
and use it for their mine planning.</p>
<p>[00:02:16.920]<br />
So I’m just going to stop my camera for now</p>
<p>[00:02:21.012]<br />
so that, there we go, turning it off and we’ll carry on.</p>
<p>[00:02:25.290]<br />
Now, this grade-thickness in Edge is one way to do it.</p>
<p>[00:02:29.820]<br />
We also have presented in the past,</p>
<p>[00:02:33.076]<br />
most recently at the Lyceum 2020 Tips &amp; Tricks</p>
<p>[00:02:37.500]<br />
with Sarah Connolly presenting</p>
<p>[00:02:40.312]<br />
and we’ll provide a link for you to this recording</p>
<p>[00:02:44.240]<br />
that was done last fall.</p>
<p>[00:02:45.990]<br />
So this is a way to do grade-thickness contouring in Geo,</p>
<p>[00:02:49.260]<br />
if you don’t have Edge.</p>
<p>[00:02:53.280]<br />
All right, now I’ll flip to the live demonstration.</p>
<p>[00:02:59.490]<br />
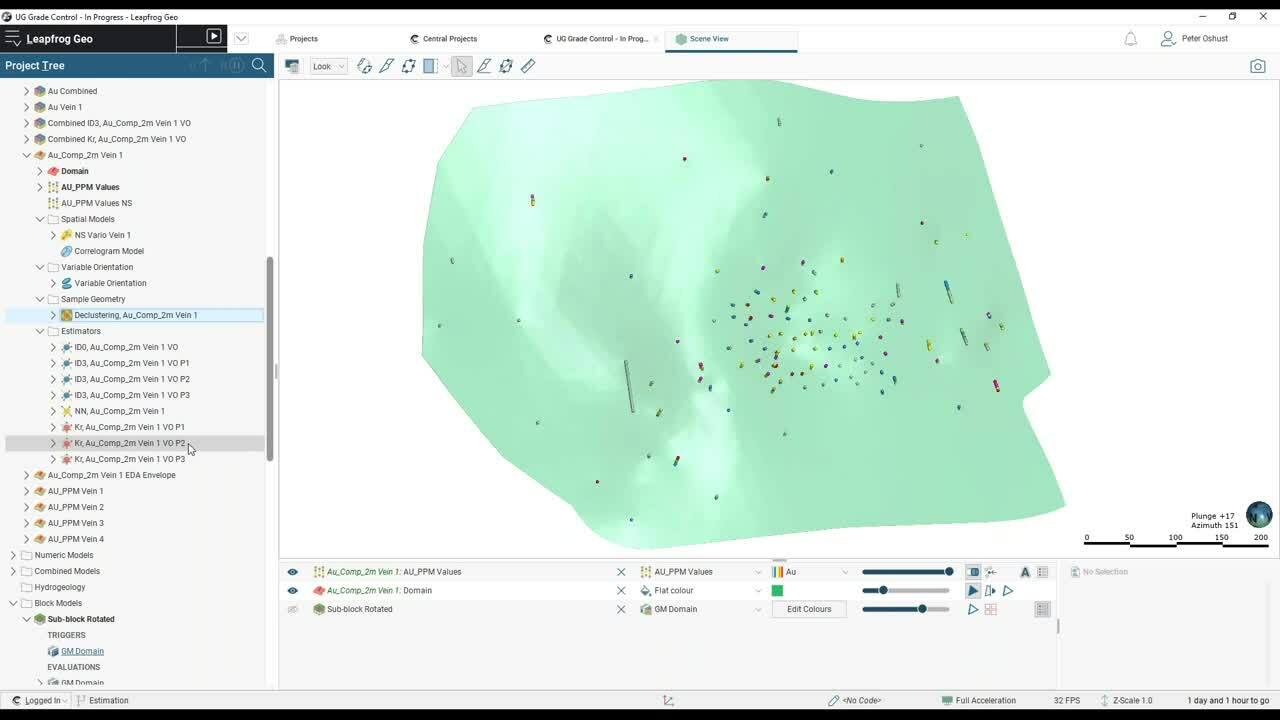
And we’re starting off with a set of veins.</p>
<p>[00:03:04.430]<br />
There are four veins here, quite a variety of drill holes,</p>
<p>[00:03:09.540]<br />
not a lot of sapling, but that’s kind of common</p>
<p>[00:03:12.480]<br />
with the many narrow vein situations</p>
<p>[00:03:14.790]<br />
because it’s difficult to reach them.</p>
<p>[00:03:17.700]<br />
Now let’s see what else we’ve got here.</p>
<p>[00:03:19.170]<br />
So that’s all of our veins and all of the drill holes.</p>
<p>[00:03:23.040]<br />
I’m going to load another scene,</p>
<p>[00:03:26.050]<br />
which will show us what we’ve got for vein 1.</p>
<p>[00:03:30.290]<br />
And it looks like I must have overwritten that scene.</p>
<p>[00:03:35.060]<br />
So I’ll just turn off some of these other veins.</p>
<p>[00:03:38.350]<br />
So here’s vein 1 with all of the drill holes.</p>
<p>[00:03:42.686]<br />
So let’s filter some of these for vein 1.</p>
<p>[00:03:47.530]<br />
So those are just the assays that we have in vein 1</p>
<p>[00:03:51.536]<br />
and it’s pretty typical that we’ve got clustered data,</p>
<p>[00:03:54.586]<br />
so they’ve really drilled this area off quite well</p>
<p>[00:03:56.280]<br />
with some scattered holes out around the edge,</p>
<p>[00:04:00.622]<br />
maybe chasing the extents of that structure.</p>
<p>[00:04:03.500]<br />
So in other words, we’ve got some clustered data here.</p>
<p>[00:04:07.380]<br />
So that’s a quick review of the data in the scene.</p>
<p>[00:04:10.830]<br />
Now I’m going to flip to just looking at the sample lengths,</p>
<p>[00:04:15.970]<br />
the assay interval lengths because those will help us</p>
<p>[00:04:19.860]<br />
in making a decision on what composite length to use.</p>
<p>[00:04:23.140]<br />
So this is the first of our EDA in the drill hole data.</p>
<p>[00:04:27.480]<br />
And I’m going to go to the original assay table.</p>
<p>[00:04:32.590]<br />
I do have a merged table that has the evaluated</p>
<p>[00:04:37.180]<br />
geological model combined with the assays,</p>
<p>[00:04:39.750]<br />
so we can do some filters on that.</p>
<p>[00:04:42.000]<br />
But let’s just check the statistics on our table.</p>
<p>[00:04:45.640]<br />
So at the table level, we have multivariate statistics</p>
<p>[00:04:49.009]<br />
and we have access to the interval lengths statistics.</p>
<p>[00:04:52.800]<br />
So looking at a cumulative histogram of sample lengths,</p>
<p>[00:04:56.720]<br />
we can see that we have about 80% of the samples</p>
<p>[00:04:59.750]<br />
were taken at one meter or less</p>
<p>[00:05:03.160]<br />
and then about 20 are higher</p>
<p>[00:05:06.510]<br />
and there’s quite a kick at two.</p>
<p>[00:05:10.286]<br />
So it looks like, well, if we look at the histogram,</p>
<p>[00:05:11.930]<br />
we’ll probably see the mode there,</p>
<p>[00:05:14.790]<br />
a big mode at one,</p>
<p>[00:05:16.560]<br />
but then it’s a little bumped down here at two meters.</p>
<p>[00:05:19.850]<br />
So given that we don’t want to subdivide</p>
<p>[00:05:23.260]<br />
our longer sample intervals,</p>
<p>[00:05:27.186]<br />
which would impact the coefficient of variation, the CV,</p>
<p>[00:05:33.310]<br />
it might make our data look a little bit better than it is,</p>
<p>[00:05:36.390]<br />
so we’ll composite to two meters.</p>
<p>[00:05:39.340]<br />
And that way we’ll get a better distribution of composites.</p>
<p>[00:05:47.110]<br />
And we are expecting to see some changes,</p>
<p>[00:05:50.020]<br />
but let’s look at what we’ve done for compositing.</p>
<p>[00:05:54.490]<br />
I have a two-meter composite table here.</p>
<p>[00:05:56.600]<br />
Just have a look at what we’ve done to composite.</p>
<p>[00:06:00.486]<br />
So I’ve composited inside the evaluated GM,</p>
<p>[00:06:03.920]<br />
that’s the back flag, if you want to think of it,</p>
<p>[00:06:05.620]<br />
the back flag models, compositing to two meters.</p>
<p>[00:06:08.950]<br />
And if we have any residual end lengths, one meter and less,</p>
<p>[00:06:14.220]<br />
they get tagged or backstitched I should say,</p>
<p>[00:06:18.030]<br />
backstitched to the previous assay intervals.</p>
<p>[00:06:20.450]<br />
So that’s how we manage that</p>
<p>[00:06:22.441]<br />
and composited all of the assay values.</p>
<p>[00:06:27.720]<br />
So we have composites.</p>
<p>[00:06:30.220]<br />
So let’s have a look at what the composites look like</p>
<p>[00:06:33.330]<br />
in the scene then, I think I’ve got a scene saved for that.</p>
<p>[00:06:38.150]<br />
No, I must have overwritten that one too.</p>
<p>[00:06:40.040]<br />
I was playing on here this morning</p>
<p>[00:06:44.251]<br />
and made a few new scenes,</p>
<p>[00:06:46.640]<br />
I must have wrecked my earlier ones.</p>
<p>[00:06:48.970]<br />
So let’s just get rid of the domain assays.</p>
<p>[00:06:54.690]<br />
Click on the right spot and then load the composite.</p>
<p>[00:06:59.130]<br />
So we have gold composites,</p>
<p>[00:07:02.990]<br />
pretty much the same distribution.</p>
<p>[00:07:06.370]<br />
And if I apply the vein 1 filter,</p>
<p>[00:07:09.080]<br />
we’re not going to see too much different either.</p>
<p>[00:07:13.240]<br />
Turn on the rendered tubes and make them fairly fat</p>
<p>[00:07:18.200]<br />
so we can see what we’ve got here.</p>
<p>[00:07:20.591]<br />
So there’s the composites.</p>
<p>[00:07:23.679]<br />
Now we do have, again, that clustered data in here</p>
<p>[00:07:27.770]<br />
and we have some areas where we had missing intervals</p>
<p>[00:07:32.840]<br />
and now those intervals have been,</p>
<p>[00:07:36.150]<br />
composite intervals have been created in there.</p>
<p>[00:07:38.140]<br />
So I think the numbers of our composites have gone up,</p>
<p>[00:07:41.450]<br />
but that’s a good thing really,</p>
<p>[00:07:43.790]<br />
because we’ve got some low values in here</p>
<p>[00:07:45.990]<br />
now that we can use to constrain the estimate.</p>
<p>[00:07:49.500]<br />
Well, let’s just check the stats on our composites.</p>
<p>[00:07:57.210]<br />
So we have 354 samples,</p>
<p>[00:07:59.890]<br />
with an average of 1.9 approximately grams</p>
<p>[00:08:04.030]<br />
and a CV of just over two.</p>
<p>[00:08:06.570]<br />
So it does have some variants in this distribution</p>
<p>[00:08:11.910]<br />
and it looks like we’ve got some outliers over here.</p>
<p>[00:08:16.690]<br />
I did an analysis previously,</p>
<p>[00:08:18.540]<br />
earlier on and I pegged 23 grams as the outliers.</p>
<p>[00:08:22.940]<br />
If I want to see where these are in the scene</p>
<p>[00:08:25.440]<br />
and you may know already that you can interact</p>
<p>[00:08:28.060]<br />
from charts in the scene, it applies a dynamic filter.</p>
<p>[00:08:32.030]<br />
So I’ve selected the tail in that histogram distribution,</p>
<p>[00:08:35.720]<br />
and it’s filtered now for those composites in the scene</p>
<p>[00:08:38.920]<br />
and I can see the distribution of them.</p>
<p>[00:08:42.299]<br />
Now, if they’re almost clustered,</p>
<p>[00:08:43.550]<br />
which means we might consider</p>
<p>[00:08:45.920]<br />
using an outlier search restriction on these,</p>
<p>[00:08:48.440]<br />
but I chose to cap them.</p>
<p>[00:08:50.680]<br />
Generally speaking, if you have a cluster of outliers,</p>
<p>[00:08:53.630]<br />
it may reflect the fact that there’s a subpopulation</p>
<p>[00:08:57.693]<br />
that hasn’t been modeled out, so you can treat it</p>
<p>[00:08:59.927]<br />
with a special outlier search restriction.</p>
<p>[00:09:05.090]<br />
I’ll just get my vein 1 filter back here.</p>
<p>[00:09:08.950]<br />
All right, so it was predetermined</p>
<p>[00:09:12.090]<br />
that we would use vein 1 as a domain estimation.</p>
<p>[00:09:14.830]<br />
So the next step in the workflow</p>
<p>[00:09:17.180]<br />
is to add a domain estimation to our Estimations folder.</p>
<p>[00:09:22.740]<br />
If you’re a Geo user, you won’t see estimations.</p>
<p>[00:09:26.710]<br />
It’s only when you have the Edge extension</p>
<p>[00:09:28.460]<br />
that you see this folder</p>
<p>[00:09:30.940]<br />
where you can define the estimations</p>
<p>[00:09:33.420]<br />
that you want to evaluate onto your block model.</p>
<p>[00:09:36.370]<br />
So let’s just clear the scene on this one.</p>
<p>[00:09:39.760]<br />
And I think I do have a scene ready to go for to this.</p>
<p>[00:09:43.980]<br />
So let’s go to saved scenes.</p>
<p>[00:09:46.070]<br />
Here’s my domain estimation using saved scenes,</p>
<p>[00:09:50.530]<br />
just saves a few clicks.</p>
<p>[00:09:53.460]<br />
Well, not a lot has changed from what we were looking at</p>
<p>[00:09:55.890]<br />
in terms of composites, but you’ll see now</p>
<p>[00:09:59.369]<br />
that where we had intervals in the drill holes before,</p>
<p>[00:10:01.820]<br />
now we have discrete points.</p>
<p>[00:10:03.640]<br />
So those reflect the centroids</p>
<p>[00:10:05.750]<br />
or the centers of the composites.</p>
<p>[00:10:10.040]<br />
And let’s have a look at, that’s the 3D view.</p>
<p>[00:10:13.910]<br />
And let’s just check that the stats still look okay.</p>
<p>[00:10:17.750]<br />
So go back up to the Estimation folder to the vein 1.</p>
<p>[00:10:23.320]<br />
I guess I can show you what the boundary looks like too.</p>
<p>[00:10:26.930]<br />
So I’m just double clicking on the domain estimation.</p>
<p>[00:10:30.930]<br />
It’s calculating a boundary block</p>
<p>[00:10:33.930]<br />
showing us the average grade inside the vein versus outside.</p>
<p>[00:10:39.170]<br />
And what we’re paying attention to here</p>
<p>[00:10:40.940]<br />
is what’s going on across the boundary.</p>
<p>[00:10:43.930]<br />
And in this case, there’s quite a sharp drop in grade.</p>
<p>[00:10:47.040]<br />
There’s a high grade contrast.</p>
<p>[00:10:49.250]<br />
So I will use a hard boundary.</p>
<p>[00:10:51.450]<br />
Now if this was more gradational,</p>
<p>[00:10:54.143]<br />
if the boundary was a little bit fuzzy, for instance,</p>
<p>[00:10:55.850]<br />
I could use a soft boundary and share samples</p>
<p>[00:10:59.300]<br />
from the outside to a specified distance.</p>
<p>[00:11:02.656]<br />
And this is calculated as perpendicular</p>
<p>[00:11:06.050]<br />
to the triangle faces, so it’s nearly a true distance.</p>
<p>[00:11:09.900]<br />
But I am going to use a hard boundary, just cancel that.</p>
<p>[00:11:14.600]<br />
And we’ll start looking at some of the other things here.</p>
<p>[00:11:17.010]<br />
So we’ve got the domain and the values loaded.</p>
<p>[00:11:19.830]<br />
I also did a normal scores transform</p>
<p>[00:11:22.240]<br />
in an effort to calculate a nicer variogram,</p>
<p>[00:11:27.030]<br />
but in the end, I didn’t go there.</p>
<p>[00:11:30.425]<br />
But we do have the ability to do</p>
<p>[00:11:32.370]<br />
a Gaussian Weierstrass</p>
<p>[00:11:34.670]<br />
or discreet Gaussian transformation,</p>
<p>[00:11:36.800]<br />
whatever you want to call it.</p>
<p>[00:11:38.840]<br />
Anyway, so it’s a normal scores transform,</p>
<p>[00:11:42.452]<br />
which can sometimes help to improve modeling,</p>
<p>[00:11:45.740]<br />
calculating and modeling variograms</p>
<p>[00:11:49.162]<br />
in the presence of noisy data.</p>
<p>[00:11:50.890]<br />
I opted instead, and we’ll go the correlogram here.</p>
<p>[00:11:54.810]<br />
So we’re looking now at our spatial continuity in our model.</p>
<p>[00:11:59.190]<br />
I opted to go with a correlogram</p>
<p>[00:12:02.130]<br />
because a correlogram will work better</p>
<p>[00:12:06.150]<br />
in the presence of clustered data.</p>
<p>[00:12:08.530]<br />
It is very good at managing to control outliers, noisy data,</p>
<p>[00:12:14.250]<br />
and it also helps to see past the clustered data.</p>
<p>[00:12:19.590]<br />
So the correlogram, it’s the covariance function,</p>
<p>[00:12:24.810]<br />
which has been normalized to the mean of the sample pairs,</p>
<p>[00:12:30.630]<br />
if you want to think of it that way.</p>
<p>[00:12:32.190]<br />
So the covariance function is divided by the,</p>
<p>[00:12:38.112]<br />
the mean squared of the data or the standard deviation</p>
<p>[00:12:41.410]<br />
of the data to normalize it to the mean.</p>
<p>[00:12:44.640]<br />
So we can see in our map</p>
<p>[00:12:47.992]<br />
that there is quite a strong vertical trend,</p>
<p>[00:12:50.810]<br />
the pitch actually is the vertical</p>
<p>[00:12:53.820]<br />
and the correlograms are displayed in their true form,</p>
<p>[00:12:57.610]<br />
which is inverted to how we usually display</p>
<p>[00:13:01.500]<br />
traditional semi-variograms.</p>
<p>[00:13:04.610]<br />
Anyway, so that’s why these curves are upside down.</p>
<p>[00:13:07.400]<br />
And I have managed to apply reasonable models, I think,</p>
<p>[00:13:11.430]<br />
to the experimental variograms, so if we look in the scene,</p>
<p>[00:13:16.213]<br />
we’ll see our variogram ellipse.</p>
<p>[00:13:20.300]<br />
It’s not very big, so the continuity isn’t great.</p>
<p>[00:13:23.530]<br />
But it does look reasonable together with the data.</p>
<p>[00:13:26.400]<br />
So that is the go ahead variogram.</p>
<p>[00:13:31.920]<br />
And the next thing we want to look at is declustering.</p>
<p>[00:13:35.460]<br />
So clustered data will give you</p>
<p>[00:13:40.500]<br />
typically an overstated naive mean of your samples.</p>
<p>[00:13:46.640]<br />
So we use different declustering methods.</p>
<p>[00:13:49.590]<br />
Leapfrog provides a moving window declustering.</p>
<p>[00:13:54.120]<br />
You can also use a nearest neighbor model,</p>
<p>[00:13:56.440]<br />
which gives you in effect like a 3D</p>
<p>[00:13:59.950]<br />
polygonal volume type of a weighting.</p>
<p>[00:14:02.615]<br />
So the samples are weighted by the inverse</p>
<p>[00:14:04.580]<br />
of the area that they have around them.</p>
<p>[00:14:09.140]<br />
And so these outline samples get more weight</p>
<p>[00:14:13.270]<br />
than the closely spaced,</p>
<p>[00:14:15.940]<br />
typically higher grade clustered data</p>
<p>[00:14:18.660]<br />
where we’ve drilled off the higher grade</p>
<p>[00:14:20.660]<br />
portion of the vein.</p>
<p>[00:14:22.980]<br />
And so the declustered mean</p>
<p>[00:14:24.930]<br />
is typically lower than your naive mean.</p>
<p>[00:14:29.190]<br />
So let’s have a look if that’s the case</p>
<p>[00:14:30.690]<br />
with our declustering tool in Edge.</p>
<p>[00:14:34.780]<br />
So here’s our distribution.</p>
<p>[00:14:38.032]<br />
And you can see as the moving window relative size</p>
<p>[00:14:41.290]<br />
increases, that the average grade comes down</p>
<p>[00:14:45.360]<br />
until it hits kind of a trough</p>
<p>[00:14:47.250]<br />
and then it goes back up again.</p>
<p>[00:14:49.922]<br />
And I suppose if you used a search ellipse</p>
<p>[00:14:52.632]<br />
that was the same size as your vein,</p>
<p>[00:14:53.530]<br />
it would come right back up to the input.</p>
<p>[00:14:56.640]<br />
So the input mean, and this mean is a little bit different</p>
<p>[00:15:00.030]<br />
than the 1.91 that we saw earlier.</p>
<p>[00:15:02.280]<br />
So this is the mean of the points,</p>
<p>[00:15:06.332]<br />
of the data rather than the length weighted intervals.</p>
<p>[00:15:08.360]<br />
So it’s just slightly different,</p>
<p>[00:15:09.450]<br />
but very close, 1.89, roughly.</p>
<p>[00:15:12.620]<br />
And if I click on this node,</p>
<p>[00:15:14.740]<br />
we’ll see that the declustered mean is 1.66.</p>
<p>[00:15:18.820]<br />
So file that number away for later.</p>
<p>[00:15:23.490]<br />
That’s our target.</p>
<p>[00:15:26.060]<br />
Moving on to estimators,</p>
<p>[00:15:29.608]<br />
and I defined three different estimators.</p>
<p>[00:15:32.890]<br />
There’s an inverse distance cubed,</p>
<p>[00:15:35.700]<br />
a nearest neighbor and a Creed estimator.</p>
<p>[00:15:40.707]<br />
And with the CV up around two,</p>
<p>[00:15:42.230]<br />
I was expecting that I would need</p>
<p>[00:15:44.000]<br />
a slightly more selective type of an estimator</p>
<p>[00:15:46.940]<br />
and that’s why IDCUBE.</p>
<p>[00:15:49.922]<br />
And we’ll see what the results are comparing the Creed</p>
<p>[00:15:54.230]<br />
to the inverse distance cubed.</p>
<p>[00:15:59.708]<br />
Now I did a multi-pass strategy as well,</p>
<p>[00:16:03.106]<br />
and that again, is to address the clustered data.</p>
<p>[00:16:06.520]<br />
So the first pass search,</p>
<p>[00:16:08.360]<br />
I’ll open up the first pass search here</p>
<p>[00:16:10.760]<br />
using the variogram and the ellipsoid</p>
<p>[00:16:13.370]<br />
is to the variogram limit.</p>
<p>[00:16:15.610]<br />
So it looked reasonable as a starting point</p>
<p>[00:16:17.810]<br />
for a first pass.</p>
<p>[00:16:20.545]<br />
And I have set a minimum number of samples of 14</p>
<p>[00:16:24.100]<br />
and a maximum of 20</p>
<p>[00:16:25.780]<br />
and a maximum samples per drill hole of five.</p>
<p>[00:16:28.750]<br />
That means that I need three holes on my first pass</p>
<p>[00:16:32.430]<br />
to estimate a block.</p>
<p>[00:16:34.780]<br />
And the block search isn’t very wide,</p>
<p>[00:16:38.490]<br />
so I’m trying to minimize the negative Creeding weights.</p>
<p>[00:16:42.390]<br />
So as the passes go, then the second pass uses a maximum</p>
<p>[00:16:48.330]<br />
or two holes to estimate the block.</p>
<p>[00:16:51.760]<br />
And then finally pass three, I’ll just show you,</p>
<p>[00:16:54.320]<br />
is pretty wide open, big search</p>
<p>[00:16:57.530]<br />
and no restrictions on the samples.</p>
<p>[00:17:00.831]<br />
So even one sample will result in a block grade estimate.</p>
<p>[00:17:04.080]<br />
So the idea here that this is just a fill pass,</p>
<p>[00:17:07.760]<br />
making sure that as many blocks as possible are estimated,</p>
<p>[00:17:10.690]<br />
and I use the similar strategies for the same sorry,</p>
<p>[00:17:15.390]<br />
same search in sample selection for the IDCUBE.</p>
<p>[00:17:19.830]<br />
So what does this look like when we evaluate it in a model?</p>
<p>[00:17:24.920]<br />
Well, I guess first off, we’ll build a model.</p>
<p>[00:17:28.080]<br />
Now I have one built already,</p>
<p>[00:17:30.322]<br />
but as I mentioned, the kind of the trick</p>
<p>[00:17:32.480]<br />
to doing the grade-thickness in Edge</p>
<p>[00:17:34.930]<br />
is to define a rotated sub-block model.</p>
<p>[00:17:37.840]<br />
So let’s see what that looks like.</p>
<p>[00:17:40.650]<br />
A new sub-block model,</p>
<p>[00:17:44.530]<br />
big parent blocks.</p>
<p>[00:17:45.530]<br />
The parent blocks are scaled to the project limits</p>
<p>[00:17:49.870]<br />
and it thinks that I need to have huge blocks</p>
<p>[00:17:53.880]<br />
because the topography is very extensive,</p>
<p>[00:17:58.522]<br />
but it’s not the case.</p>
<p>[00:17:59.420]<br />
Now I’m going to use a 10 by 10 in the X and the Y</p>
<p>[00:18:03.240]<br />
and the 300 in Z and when I’m done,</p>
<p>[00:18:06.450]<br />
I will have the, well, the next step actually</p>
<p>[00:18:08.810]<br />
is to rotate the model such that Z is across the vein</p>
<p>[00:18:13.950]<br />
and that way, with a variable Z</p>
<p>[00:18:16.540]<br />
going from zero to whatever it needs to be,</p>
<p>[00:18:19.440]<br />
it will make like an array of blocks</p>
<p>[00:18:21.920]<br />
that kind of look like little prisms.</p>
<p>[00:18:25.231]<br />
There’s a shortcut to set the angles of a rotated model,</p>
<p>[00:18:29.070]<br />
and that is to use a trend plane.</p>
<p>[00:18:30.860]<br />
So let’s just go back to the scene,</p>
<p>[00:18:33.370]<br />
align the camera up to look down dip of that vein</p>
<p>[00:18:39.542]<br />
and I’ll just apply a trend plane here.</p>
<p>[00:18:42.280]<br />
That’s got to be about 305.</p>
<p>[00:18:45.940]<br />
I’m just going to edit this,</p>
<p>[00:18:47.795]<br />
305 and the dip, 66 is okay.</p>
<p>[00:18:51.090]<br />
I don’t need to worry about the pitch for this step.</p>
<p>[00:18:54.600]<br />
It doesn’t come to bear</p>
<p>[00:18:56.730]<br />
when I’m defining the block model geometry.</p>
<p>[00:18:59.460]<br />
So now, I will set my angles from the moving plane.</p>
<p>[00:19:03.930]<br />
And now that model, it’s a little bit jumped off</p>
<p>[00:19:08.900]<br />
to the side there.</p>
<p>[00:19:11.045]<br />
It is in the correct orientation now</p>
<p>[00:19:15.500]<br />
at least for that vein, where am I?</p>
<p>[00:19:21.230]<br />
Oh, my trend plane isn’t very good.</p>
<p>[00:19:23.000]<br />
Let’s back up here.</p>
<p>[00:19:24.300]<br />
I’m going to define a trend plane first</p>
<p>[00:19:31.382]<br />
and then it’ll create a plane.</p>
<p>[00:19:34.292]<br />
To the side, 305</p>
<p>[00:19:37.140]<br />
and dipping, 66 I think was good enough.</p>
<p>[00:19:42.360]<br />
Now a trend plane and let’s get back</p>
<p>[00:19:44.905]<br />
to this block model business.</p>
<p>[00:19:46.380]<br />
New sub-block model,</p>
<p>[00:19:50.402]<br />
10 by 10 by 300.</p>
<p>[00:19:52.710]<br />
I want to make sure that I go right across the vein</p>
<p>[00:19:55.410]<br />
wherever there are any undulations.</p>
<p>[00:19:58.180]<br />
And I will just have five-meter sub-blocks.</p>
<p>[00:20:01.490]<br />
So the sub-block count two into 10</p>
<p>[00:20:05.070]<br />
gives me my two five-meter sub-blocks</p>
<p>[00:20:09.830]<br />
and let’s set angles from moving plane.</p>
<p>[00:20:15.890]<br />
It’s better.</p>
<p>[00:20:16.780]<br />
Now it’s lined up to where that vein is.</p>
<p>[00:20:19.455]<br />
There’s a lot of extra real estate here that we can get.</p>
<p>[00:20:24.480]<br />
We can trim that by moving the extents</p>
<p>[00:20:27.940]<br />
a little bit back and forth.</p>
<p>[00:20:30.928]<br />
Of course, the minimum thickness of that model</p>
<p>[00:20:34.030]<br />
in the Z direction is going to be 300</p>
<p>[00:20:36.370]<br />
because that’s what I have set my Z dimension to be.</p>
<p>[00:20:43.530]<br />
One more tweak and that’s roughed in pretty, pretty good.</p>
<p>[00:20:50.640]<br />
Of course, if you had an open pit,</p>
<p>[00:20:51.990]<br />
you would have to make it a little bit bigger,</p>
<p>[00:20:54.630]<br />
but this is going to be an underground mine.</p>
<p>[00:20:57.270]<br />
So that’s aligning it to the model</p>
<p>[00:21:00.830]<br />
and you can see by the checkerboard pattern</p>
<p>[00:21:03.090]<br />
that Z is across the vein.</p>
<p>[00:21:06.327]<br />
And then after that, I would set my sub-blocking triggers</p>
<p>[00:21:07.960]<br />
and devaluations and carry on.</p>
<p>[00:21:10.670]<br />
Now we already have a model built.</p>
<p>[00:21:12.410]<br />
So I’ll just click Cancel at this point</p>
<p>[00:21:16.735]<br />
and bring that model into the scene.</p>
<p>[00:21:19.805]<br />
Well, the first thing we could look at</p>
<p>[00:21:21.720]<br />
is the evaluated geological model.</p>
<p>[00:21:25.010]<br />
Oops, the evaluated geological model</p>
<p>[00:21:29.070]<br />
and that is filtered for measured and indicated blocks,</p>
<p>[00:21:31.940]<br />
but there’s the model.</p>
<p>[00:21:35.739]<br />
And if we cut a slice right along the model trend</p>
<p>[00:21:41.060]<br />
slice and cross the vein</p>
<p>[00:21:47.700]<br />
and then set the width to five and the step size to five.</p>
<p>[00:21:52.770]<br />
And this can be 125.</p>
<p>[00:21:55.820]<br />
Now I am looking perpendicular to the model.</p>
<p>[00:22:00.440]<br />
And if I hit L, on the keyboard to look at that model,</p>
<p>[00:22:07.040]<br />
I should be able to see those prisms</p>
<p>[00:22:09.840]<br />
that I was looking for.</p>
<p>[00:22:10.890]<br />
Yeah, they’re all kind of prisms.</p>
<p>[00:22:12.540]<br />
We can see this model isn’t very thick or tall,</p>
<p>[00:22:15.280]<br />
it’s only about five meters or less.</p>
<p>[00:22:19.940]<br />
And I don’t see any breaks in the block.</p>
<p>[00:22:22.290]<br />
So that means that my Z value at 300 is pretty good.</p>
<p>[00:22:26.260]<br />
If I would have used a Z at, let’s say a 100 meters,</p>
<p>[00:22:29.780]<br />
I may have had some blocks being split,</p>
<p>[00:22:32.300]<br />
but I want only blocks that are completely across</p>
<p>[00:22:36.454]<br />
that vein in the Z direction.</p>
<p>[00:22:40.960]<br />
So let’s turn off these lights, we’ve got our model</p>
<p>[00:22:44.360]<br />
and of course we’ve evaluated the GM</p>
<p>[00:22:47.560]<br />
and I set the sub-blocking triggers to the GM as well.</p>
<p>[00:22:51.890]<br />
Now I’m just going to my cheat sheet here</p>
<p>[00:22:53.750]<br />
to see what I also want to show you.</p>
<p>[00:22:56.580]<br />
So at this point, yeah,</p>
<p>[00:22:58.239]<br />
let’s have a quick look at some of the models.</p>
<p>[00:23:00.560]<br />
So that’s the geology.</p>
<p>[00:23:04.060]<br />
I evaluated,</p>
<p>[00:23:05.360]<br />
I created, sorry, I created a combined estimator</p>
<p>[00:23:08.170]<br />
for the three passes for the inverse distance cubed</p>
<p>[00:23:12.040]<br />
and the Creed estimator</p>
<p>[00:23:13.900]<br />
so that I can combine all three passes.</p>
<p>[00:23:16.810]<br />
Actually, I better show you what that looks like.</p>
<p>[00:23:19.180]<br />
So in the combined estimator,</p>
<p>[00:23:21.410]<br />
I just double clicked on it to open.</p>
<p>[00:23:23.320]<br />
I selected passes one, two, and three in order.</p>
<p>[00:23:26.528]<br />
It’s important because as a block is estimated,</p>
<p>[00:23:30.930]<br />
it doesn’t get overwritten by the following passes.</p>
<p>[00:23:34.700]<br />
So if I were to put pass three at the top,</p>
<p>[00:23:37.160]<br />
of course, everything would have been estimated</p>
<p>[00:23:38.680]<br />
with pass three and pass one and two</p>
<p>[00:23:40.610]<br />
wouldn’t have had any impact on the model at all.</p>
<p>[00:23:43.130]<br />
So yes, hierarchy is important and it is correct.</p>
<p>[00:23:48.210]<br />
So let’s just have a look at that model.</p>
<p>[00:23:51.690]<br />
So there’s the Creed, here’s the Creed model</p>
<p>[00:23:55.745]<br />
and it’s not bad, but I can see,</p>
<p>[00:23:59.420]<br />
I would have to tune it a little bit better.</p>
<p>[00:24:01.410]<br />
I think there’s some funny artifact patches of blocks</p>
<p>[00:24:04.870]<br />
and things, may or may not be able to get rid of those.</p>
<p>[00:24:11.520]<br />
And there are big areas around the edge</p>
<p>[00:24:15.370]<br />
that have just one grade.</p>
<p>[00:24:17.480]<br />
So that’s kind of reflecting</p>
<p>[00:24:19.518]<br />
the fact that there’s not a lot of data out there</p>
<p>[00:24:21.180]<br />
and that third pass search basically estimating the block</p>
<p>[00:24:24.490]<br />
with that one pass.</p>
<p>[00:24:26.670]<br />
Let’s see what the IDCUBE model looks like.</p>
<p>[00:24:31.000]<br />
That’s a much prettier model, I guess</p>
<p>[00:24:33.430]<br />
because the thing is how does it validate?</p>
<p>[00:24:37.195]<br />
And we will check it against the nearest neighbor model,</p>
<p>[00:24:40.750]<br />
which is kind of a proxy to a declustered distribution.</p>
<p>[00:24:44.150]<br />
Even though we will have more than one sample per block,</p>
<p>[00:24:48.280]<br />
it does kind of emulate or is a proxy for</p>
<p>[00:24:52.343]<br />
properly declustered to distribution.</p>
<p>[00:24:55.990]<br />
Anyway, let’s go back to the IDCUBE.</p>
<p>[00:24:59.260]<br />
Another thing that we can do,</p>
<p>[00:25:01.330]<br />
and that is to restrict our comparison</p>
<p>[00:25:06.490]<br />
within a reasonable envelope</p>
<p>[00:25:08.870]<br />
around the blocks that are well-supported.</p>
<p>[00:25:14.042]<br />
So this is basically, where does it matter?</p>
<p>[00:25:15.980]<br />
Like it doesn’t matter so much around the edges,</p>
<p>[00:25:18.580]<br />
we’re expecting a little bit of error out there.</p>
<p>[00:25:24.795]<br />
But if we define a boundary</p>
<p>[00:25:27.010]<br />
around that are of the well-drilled region,</p>
<p>[00:25:35.360]<br />
and it’s showing in there.</p>
<p>[00:25:38.190]<br />
There’s my well-drilled region.</p>
<p>[00:25:39.550]<br />
So I’m calling this my EDA envelope.</p>
<p>[00:25:42.050]<br />
I’m going to do my validation checks in that envelope.</p>
<p>[00:25:45.955]<br />
They’re going to be much more relevant</p>
<p>[00:25:48.745]<br />
than just having everything on the outside</p>
<p>[00:25:51.870]<br />
that is inferred confidence or less, let’s say.</p>
<p>[00:25:55.720]<br />
Okay, back to the model and let’s check our stats.</p>
<p>[00:26:01.090]<br />
So going to check statistics, table of statistics.</p>
<p>[00:26:07.505]<br />
And I want to replicate the mean of the distribution</p>
<p>[00:26:13.500]<br />
with my estimates.</p>
<p>[00:26:14.920]<br />
And you’ll recall that the declustered mean is 1.66,</p>
<p>[00:26:19.845]<br />
the nearest neighbor model is also very close to that</p>
<p>[00:26:22.600]<br />
within a percent, 1.689, and the CV is almost the same</p>
<p>[00:26:28.240]<br />
as it was for our samples, which was two.</p>
<p>[00:26:30.982]<br />
So that nearest neighbor model,</p>
<p>[00:26:33.116]<br />
again, reasonable proxy</p>
<p>[00:26:35.550]<br />
for the declustered grade distribution</p>
<p>[00:26:37.970]<br />
and very useful for comparison.</p>
<p>[00:26:40.040]<br />
The IDCUBE model comes in quite well.</p>
<p>[00:26:43.960]<br />
Well, it’s a little bit off, but not bad, 1.73, roughly.</p>
<p>[00:26:47.400]<br />
So we’re replicating the mean of our input distribution</p>
<p>[00:26:52.530]<br />
with the IDCUBE.</p>
<p>[00:26:54.530]<br />
For some reason, we’ve got higher grades</p>
<p>[00:26:57.470]<br />
in the ordinary Creed model than we do in our samples.</p>
<p>[00:27:02.450]<br />
So that’s kind of a warning sign</p>
<p>[00:27:05.000]<br />
and it is very much smoothed,</p>
<p>[00:27:06.810]<br />
it’s .65, a CV of .66, which is much less than two,</p>
<p>[00:27:11.800]<br />
so it’s probably overly smoothed.</p>
<p>[00:27:14.290]<br />
And without doing any additional validation checks,</p>
<p>[00:27:17.280]<br />
I’m going to use my IDCUBE as the go-ahead model</p>
<p>[00:27:21.160]<br />
and yeah, carry on from there.</p>
<p>[00:27:26.240]<br />
Now let’s see.</p>
<p>[00:27:27.220]<br />
Oh, I didn’t mention it,</p>
<p>[00:27:28.932]<br />
but yeah, I did do variable orientation.</p>
<p>[00:27:31.810]<br />
Funny how you can miss stuff when you’re doing these demos.</p>
<p>[00:27:36.270]<br />
I did do a variable orientation,</p>
<p>[00:27:38.730]<br />
which is using the vein to capture the dynamic</p>
<p>[00:27:44.720]<br />
or locally varying iroinite in the estimation,</p>
<p>[00:27:49.440]<br />
our implementation of variable orientation in Edge</p>
<p>[00:27:53.450]<br />
changes the direction of the search</p>
<p>[00:27:55.980]<br />
and the direction of the variogram,</p>
<p>[00:27:57.480]<br />
it doesn’t recalculate the variogram.</p>
<p>[00:27:59.200]<br />
It just changes the directions</p>
<p>[00:28:01.120]<br />
and applies that to the search</p>
<p>[00:28:02.980]<br />
so that we get a much better local estimate</p>
<p>[00:28:06.750]<br />
using the variable orientation.</p>
<p>[00:28:09.400]<br />
Okay, so moving right along,</p>
<p>[00:28:12.818]<br />
and the next thing is to get into the calculations</p>
<p>[00:28:15.600]<br />
because that’s where we do the grade-thickness.</p>
<p>[00:28:18.390]<br />
So I’m going to double-click on Calculations,</p>
<p>[00:28:21.030]<br />
it’ll open up my calculations editor.</p>
<p>[00:28:23.540]<br />
I better show you the</p>
<p>[00:28:29.910]<br />
panel with the tools</p>
<p>[00:28:32.600]<br />
Where are my tools?</p>
<p>[00:28:36.660]<br />
I’ll maybe open it in another way here.</p>
<p>[00:28:41.455]<br />
‘Cause I have to show you that panel.</p>
<p>[00:28:42.950]<br />
Calculations and filters</p>
<p>[00:28:53.082]<br />
so there should be a panel that pops out here</p>
<p>[00:28:56.280]<br />
that we can see the metadata that is used</p>
<p>[00:29:01.450]<br />
or the items we can select, go into the calculations</p>
<p>[00:29:04.450]<br />
and our syntax buttons.</p>
<p>[00:29:08.342]<br />
And isn’t that funny?</p>
<p>[00:29:09.953]<br />
It’s not being active for me.</p>
<p>[00:29:10.950]<br />
Well, let’s have a look at the calculations anyway,</p>
<p>[00:29:12.770]<br />
because the syntax is sort of self-explanatory.</p>
<p>[00:29:16.670]<br />
So I did do a filter for the,</p>
<p>[00:29:21.010]<br />
let’s expand, collapse that.</p>
<p>[00:29:23.092]<br />
So I did do a filter for my measured in indicated,</p>
<p>[00:29:25.970]<br />
which is within the EDA envelope.</p>
<p>[00:29:28.920]<br />
So that was my limits.</p>
<p>[00:29:31.360]<br />
I also did some error traps that found empty blocks</p>
<p>[00:29:37.470]<br />
and put in a background value.</p>
<p>[00:29:39.140]<br />
They didn’t get estimated.</p>
<p>[00:29:41.055]<br />
So if it’s the vein and the estimate is normal,</p>
<p>[00:29:44.320]<br />
then it gets that value,</p>
<p>[00:29:45.620]<br />
otherwise it gets a low background value.</p>
<p>[00:29:48.030]<br />
And I did that for each of my models.</p>
<p>[00:29:50.850]<br />
I also calculated a class, category calculation for class.</p>
<p>[00:29:56.450]<br />
So if it was in the vein and it was in the EDA envelope,</p>
<p>[00:30:00.610]<br />
and within 25 meters, then it gets measured,</p>
<p>[00:30:02.940]<br />
otherwise in the EDA envelope, it’s indicated.</p>
<p>[00:30:06.090]<br />
So I did contour</p>
<p>[00:30:09.990]<br />
the region of 25 to 45 meters</p>
<p>[00:30:14.860]<br />
and then drew a poly line.</p>
<p>[00:30:17.110]<br />
And that was what formed my EDA envelope.</p>
<p>[00:30:20.010]<br />
And then outside of that, if it’s inferred</p>
<p>[00:30:23.310]<br />
or if it’s still in the vein</p>
<p>[00:30:25.030]<br />
but beyond the indicated boundary,</p>
<p>[00:30:28.677]<br />
or my EDA envelope, then I just called it inferred.</p>
<p>[00:30:33.670]<br />
So there’s also, I did for the statistics,</p>
<p>[00:30:39.390]<br />
I did also create calculated,</p>
<p>[00:30:44.360]<br />
numeric calculation of the measured and indicated blocks,</p>
<p>[00:30:49.060]<br />
two those were just for more comparisons.</p>
<p>[00:30:51.900]<br />
But finally, finally, we’re getting to the thickness.</p>
<p>[00:30:55.490]<br />
So the thickness is pretty straightforward</p>
<p>[00:30:57.050]<br />
because we have access to the Z dimension.</p>
<p>[00:31:01.000]<br />
So all I had to do for thickness</p>
<p>[00:31:03.230]<br />
is say, if it was in the vein,</p>
<p>[00:31:05.230]<br />
then give the thickness model the value of the Z dimension,</p>
<p>[00:31:09.530]<br />
otherwise it’s outside.</p>
<p>[00:31:11.620]<br />
And then the next step after that</p>
<p>[00:31:14.350]<br />
is to do a calculation, very simple.</p>
<p>[00:31:18.170]<br />
If that block was normally estimated has a value,</p>
<p>[00:31:20.780]<br />
in other words, then we just multiply our thickness</p>
<p>[00:31:24.230]<br />
times the grade of that final model.</p>
<p>[00:31:26.330]<br />
And I used the IDCUBE model, and that’s that.</p>
<p>[00:31:30.170]<br />
So we can then look at these models in the scene.</p>
<p>[00:31:37.100]<br />
Any calculation that we do can be visualized in the scene.</p>
<p>[00:31:40.460]<br />
So let’s have a look there, see the thickness,</p>
<p>[00:31:42.820]<br />
so you can see where there might be some shoots</p>
<p>[00:31:47.620]<br />
in that kind of orientation.</p>
<p>[00:31:51.435]<br />
And if we multiply, sorry, grade times thickness,</p>
<p>[00:31:56.400]<br />
we can see, yeah, maybe there are some shoots</p>
<p>[00:31:59.280]<br />
that we need to pay attention to,</p>
<p>[00:32:01.870]<br />
maybe target some holes down plunge of these shoots</p>
<p>[00:32:06.530]<br />
to see exactly what’s going on.</p>
<p>[00:32:10.084]<br />
And as I mentioned, that model exists,</p>
<p>[00:32:14.280]<br />
well exists, we can now export that model</p>
<p>[00:32:18.780]<br />
to give it to the engineers.</p>
<p>[00:32:21.490]<br />
So let’s just go to our model.</p>
<p>[00:32:24.500]<br />
What does that look like?</p>
<p>[00:32:26.350]<br />
Export,</p>
<p>[00:32:29.260]<br />
let’s call it Sub-block Model Rotated,</p>
<p>[00:32:33.550]<br />
and we can export just the CSV file</p>
<p>[00:32:36.480]<br />
that has all of the information in top of the file,</p>
<p>[00:32:42.740]<br />
a CSV with a separate text file for that metadata,</p>
<p>[00:32:46.110]<br />
or just points if you just need the points</p>
<p>[00:32:48.460]<br />
for maybe contouring or something,</p>
<p>[00:32:51.010]<br />
but I’m going to select that CV output format.</p>
<p>[00:32:56.037]<br />
Well, I’ve already done this, so it is somewhat persistent.</p>
<p>[00:32:59.670]<br />
It remembered which models I had exported</p>
<p>[00:33:03.500]<br />
and then applying a query filters</p>
<p>[00:33:06.210]<br />
so I’m not exporting the entire model,</p>
<p>[00:33:08.650]<br />
just the one for vein 1</p>
<p>[00:33:10.910]<br />
and ignoring rows or columns</p>
<p>[00:33:14.730]<br />
where all of the blocks were in air condition or empty.</p>
<p>[00:33:18.170]<br />
And then I can use status codes, either his texts</p>
<p>[00:33:22.135]<br />
or his numerics, carry on here</p>
<p>[00:33:26.100]<br />
and pick the character set.</p>
<p>[00:33:27.260]<br />
The default usually works here in North America,</p>
<p>[00:33:31.687]<br />
and there’s a summary and finally export.</p>
<p>[00:33:33.440]<br />
There aren’t a lot of blocks there,</p>
<p>[00:33:34.570]<br />
so the export actually happens pretty quickly.</p>
<p>[00:33:37.970]<br />
So let me see what else I’ve got here.</p>
<p>[00:33:44.130]<br />
Yes, okay, so the filter,</p>
<p>[00:33:46.417]<br />
I just want to show you in the block model,</p>
<p>[00:33:48.290]<br />
I did define that filter for the vein</p>
<p>[00:33:50.980]<br />
1 measured and indicated.</p>
<p>[00:33:53.250]<br />
So that is kind of the view again,</p>
<p>[00:33:56.400]<br />
where the blocks are filtered for what matters.</p>
<p>[00:34:00.210]<br />
And that’s actually is, that’s the workflow.</p>
<p>[00:34:03.740]<br />
And I hope I’ve covered it in 30 minutes or less,</p>
<p>[00:34:07.970]<br />
and the floor is now open for questions.</p>
<p>[00:34:13.010]<br />
<encoded_tag_open />v Hannah<encoded_tag_closed />Awesome, thanks, Peter.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:34:13.843]<br />
That was really good.</p>
<p>[00:34:15.670]<br />
I even learned a couple of things.</p>
<p>[00:34:18.504]<br />
I love when you sprinkle breadcrumbs of knowledge</p>
<p>[00:34:22.050]<br />
throughout your demos.</p>
<p>[00:34:24.240]<br />
Right, so we’ve got some time for questions here.</p>
<p>[00:34:26.220]<br />
I’ll give everybody a moment to type some things</p>
<p>[00:34:30.530]<br />
into that questions panel, if you haven’t done so already.</p>
<p>[00:34:35.010]<br />
I’ll start, Peter, there’s a couple of questions here.</p>
<p>[00:34:39.267]<br />
So the first one,</p>
<p>[00:34:41.217]<br />
how can you view sample distance on a block model?</p>
<p>[00:34:46.480]<br />
<encoded_tag_open />v Peter<encoded_tag_closed />Okay, maybe I’ll go back to Leapfrog<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:34:49.850]<br />
for that then.</p>
<p>[00:34:51.330]<br />
So sample or average distance and minimum distance</p>
<p>[00:34:56.000]<br />
are captured in the estimator.</p>
<p>[00:34:57.830]<br />
So let’s just go up to an estimator.</p>
<p>[00:35:07.217]<br />
Estimator, I’ll use the combined one.</p>
<p>[00:35:09.314]<br />
And in the outputs tab, if I want to see the minimum</p>
<p>[00:35:11.410]<br />
or average distance, I can select those</p>
<p>[00:35:14.140]<br />
as the output number of samples as well.</p>
<p>[00:35:18.354]<br />
So with that one selected,</p>
<p>[00:35:20.104]<br />
I should be able to go to my evaluated model.</p>
<p>[00:35:24.660]<br />
There’s my combined ID3 estimator, there’s average distance.</p>
<p>[00:35:29.637]<br />
So there’s a map of distance, to samples</p>
<p>[00:35:35.690]<br />
and each block, if I click on a block,</p>
<p>[00:35:38.070]<br />
I can actually go right to the absolute value.</p>
<p>[00:35:42.060]<br />
And you can export this too if you need that kind of thing</p>
<p>[00:35:46.400]<br />
in the exported block model.</p>
<p>[00:35:49.850]<br />
<encoded_tag_open />v Hannah<encoded_tag_closed />Okay, thanks, Peter.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:35:52.140]<br />
Another question,</p>
<p>[00:35:53.210]<br />
how can I find that grade-thickness workflow</p>
<p>[00:35:57.900]<br />
on drill holes?</p>
<p>[00:36:00.150]<br />
I can actually just paste that into the chat here.</p>
<p>[00:36:02.800]<br />
I’ll paste that hyperlink.</p>
<p>[00:36:05.910]<br />
So that was our, we had a Tips &amp; Tricks session</p>
<p>[00:36:08.150]<br />
as part of our Lyceum. I’ll put that in the chat here.</p>
<p>[00:36:12.660]<br />
Okay, another question,</p>
<p>[00:36:13.780]<br />
we saw you pick your declustering distance in the plot.</p>
<p>[00:36:20.501]<br />
Is this typically how all</p>
<p>[00:36:21.520]<br />
<encoded_tag_open />v Peter<encoded_tag_closed />I missed the question Hannah.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:36:23.414]<br />
<encoded_tag_open />v Hannah<encoded_tag_closed />We saw you pick your declustering distances<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:36:27.120]<br />
or distance in the plot, is that typically how</p>
<p>[00:36:30.010]<br />
all declustering distances are selected</p>
<p>[00:36:32.100]<br />
or can you speak more about declustering distances?</p>
<p>[00:36:36.400]<br />
<encoded_tag_open />v Peter<encoded_tag_closed />Well, generally speaking,<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:36:39.080]<br />
when we’re doing declustering,</p>
<p>[00:36:40.760]<br />
we’re targeting distribution</p>
<p>[00:36:44.750]<br />
where the area of interest has been drilled off more</p>
<p>[00:36:49.537]<br />
than outside and consequently,</p>
<p>[00:36:52.120]<br />
the naive average is higher than the declustered average.</p>
<p>[00:36:57.410]<br />
So that’s why I’m picking the lowest point here,</p>
<p>[00:37:01.940]<br />
the lowest mean from the moving window relative size.</p>
<p>[00:37:08.920]<br />
And so it’s,</p>
<p>[00:37:12.464]<br />
now that isn’t always the case.</p>
<p>[00:37:13.490]<br />
It could be flipped if you’re dealing with contaminants.</p>
<p>[00:37:16.250]<br />
In that case, you might find that your curve is upside down</p>
<p>[00:37:20.610]<br />
or inverted with respect to this one,</p>
<p>[00:37:22.680]<br />
and you would pick the highest one.</p>
<p>[00:37:25.080]<br />
So I have seen that a couple of times.</p>
<p>[00:37:26.880]<br />
I don’t have a dataset that I can emulate that,</p>
<p>[00:37:31.514]<br />
but this is typically where you’re picking</p>
<p>[00:37:34.735]<br />
your decluttering mean.</p>
<p>[00:37:36.160]<br />
Did that answer the question?</p>
<p>[00:37:37.840]<br />
<encoded_tag_open />v Hannah<encoded_tag_closed />I think so, yeah.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:37:38.910]<br />
<encoded_tag_open />v Peter<encoded_tag_closed />Okay.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:37:40.426]<br />
<encoded_tag_open />v Hannah<encoded_tag_closed />Another question just came in.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:37:42.330]<br />
I know we’re past our time here,</p>
<p>[00:37:44.170]<br />
but I do want to squeeze these out</p>
<p>[00:37:46.210]<br />
for anyone who’s interested so.</p>
<p>[00:37:47.770]<br />
Thanks, great presentation.</p>
<p>[00:37:49.370]<br />
Is there a limitation to the model size</p>
<p>[00:37:51.980]<br />
for import or export using Edge?</p>
<p>[00:37:55.887]<br />
I guess we mean block model there.</p>
<p>[00:37:57.410]<br />
<encoded_tag_open />v Peter<encoded_tag_closed />Yeah, there doesn’t appear to be a hard limit<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:38:01.987]<br />
on most block sizes.</p>
<p>[00:38:04.010]<br />
However, I should qualify that the current structure</p>
<p>[00:38:07.875]<br />
for the Edge block model does not support</p>
<p>[00:38:11.120]<br />
the importing of sub-block models.</p>
<p>[00:38:13.980]<br />
So while you can export a sub-block model,</p>
<p>[00:38:15.980]<br />
you can’t import one, which is a bit of a limitation</p>
<p>[00:38:18.660]<br />
until we fully implement the octree structure,</p>
<p>[00:38:22.210]<br />
which is similar to some of what our competitors</p>
<p>[00:38:26.727]<br />
that use sub-blocked models as well.</p>
<p>[00:38:29.827]<br />
But I know there are people out there</p>
<p>[00:38:31.850]<br />
that have billion blocked block models</p>
<p>[00:38:35.277]<br />
that they’re working actively within their organizations,</p>
<p>[00:38:38.740]<br />
mind you they’re very cumbersome at that scale.</p>
<p>[00:38:43.490]<br />
<encoded_tag_open />v Hannah<encoded_tag_closed />Right, okay.<encoded_tag_open />/v<encoded_tag_closed /></p>
<p>[00:38:47.090]<br />
Well, that wraps up our questions.</p>
<p>[00:38:50.480]<br />
We’ve got another one who says thank you.</p>
<p>[00:38:52.787]<br />
You’re welcome.</p>
<p>[00:38:53.620]<br />
So thanks again, Peter.</p>
<p>[00:38:54.825]<br />
That was awesome.</p>
<wpml_invalid_tag original=»PHA+» />[00:38:56.614]<br />
(gentle music)